2.南京医科大学公共卫生学院分子毒理学实验室;
3. 南京医科大学第一附属医院普外科
2. Department of Molecular and Genetic Toxicology, The Key Laboratory of Modern Toxicology of Ministry of Education,School of Public Health,Nanjing Medical University;
3.Department of General Surgery, The First Affiliated Hospital of Nanjing Medical University
结直肠癌是常见的恶性肿瘤之一。在全世界 范围内,其病死率位居所有恶性肿瘤死因的第二 位[1]。近年来,我国的结直肠癌发病率及死亡率也 在不断攀升。性别、年龄、饮食习惯、体重指数 (BMI)和基因等皆是影响结直肠癌发病的重要因素 [2]。最近一些研究[3]指出,miRNA的变化对结直肠癌 的发生、发展、预后等起着重要的作用。
miRNA是一类进化上保守、长度为21~23 nt 的非编码单链小RNA。它们能抑制靶基因的转 录,阻断靶蛋白的合成。已有许多研究显示, miRNA在肿瘤的发生过程中起着重要的作用。 miRNA家族中有一部分可以促进肿瘤的生长,有 一部分则会抑制肿瘤的生长[4, 5]。同时,在miRNA 的基因及其靶基因中,还存在着许多单核苷酸多态 性(SNPs)。这些多态性可能会造成miRNA的表 达、结构及功能发生变化,从而影响各种肿瘤的 发病过程。因此,这些多态性或许可以成为预测 肿瘤发生、发展及预后的生物标志[6]。
目前,国外有部分针对miRNA基因多态性与 结直肠癌发病率关系的研究,但国内对此方面的 研究较少,为此我们对结直肠癌组织中miRNA表 达谱的改变、miRNA基因及其靶基因多态性与结 直肠癌的关系及miRNA基因多态性对于结直肠癌 的诊断、预后判断、预测疗效的应用前景等内容 进行了综述。 1 结直肠癌中miRNA表达谱的改变
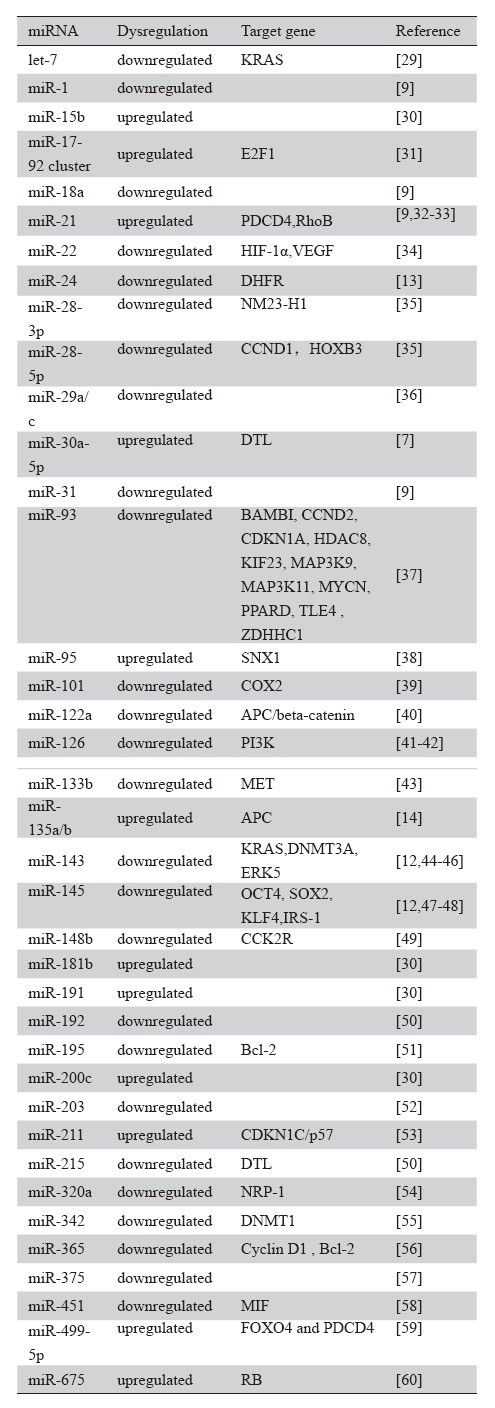
许多文献均指出,结直肠癌组织与正常组织相比,其miRNA表达水平会发生改变[7, 8, 9]。表1中 列出了部分与结直肠癌相关的miRNA及其对应的 靶基因。一些miRNA已经被发现可能具有促进或 抑制结直肠癌的作用。例如,miR-21是一种具有 致癌作用的miRNA,在进展期结肠癌组织中过表 达[8]。另一方面,一些miRNA,例如miR-1、miR- 30a-5p等,已经被证实在结直肠癌中有表达降低的 情况,因而被认为是具有抑癌作用的miRNA[7, 9]。 其他在结直肠癌中比较重要的miRNA还有miR-17– 92簇、miR-24、miR-31、miR-135a/b、miR-143、 miR-145和miR-200c等[10, 11, 12, 13, 14, 15]。
|
|
表1 结直肠癌中miRNA表达谱的改变情况 Table 1 Alterations of miRNAs in Colorectal cancer |
此外,Kyung等[19]研究了韩国人群中miR- 146a基因rs2910164位点C>G突变,miR-149基因 rs2292832位点C>T突变和miR- 499基因rs3746444 位点A>G突变的多态性与结直肠癌发病率的关 系。这些miRNA均与细胞周期和细胞凋亡的 调控机制有关。该研究的最终结果显示,miR- 146a基因rs2910164位点C>G突变和miR-149基因 rs2292832位点C>T突变的多态性在部分亚组中有 统计学意义。
|
|
表2 部分miRNA多态性与结直肠癌的关系 Table 2 Relation between miRNA polymorphism and colorectal cancer |
综合以上研究结果,我们认为部分miRNA基 因的多态性可能通过影响其靶基因的功能,进而 影响结直肠癌的发病风险。 2.2 miRNA基因多态性对于判断结直肠癌药物治疗疗效及预后的潜在价值 miRNA基因多态性也被发现与结直肠癌的药 物治疗敏感度相关。Boni等[23]检测了接受5-Fu和 CPT-11化疗的61位患者体内的18个多态性位点。 研究结果显示pri-miR26a-1的rs7372209位点多态性与肿瘤对化疗的反应和疾病进展时间有着显著 的相关性。该位点基因型为C/C和C/T的患者的治 疗效果要明显好于T/T型。此外,pri-miR-100的 rs1834306位点多态性与相对较长的疾病进展期相 关,exportin-5的rs11077位点多态性与疾病控制率 相关等也已被证实。该研究的结果说明了miRNA 生物合成系统中的多态性与结直肠癌药物治疗的 效果相关,并提示了miRNA多态性或许可以作为 接受5-Fu和CPT-11治疗的转移性结直肠癌患者临 床治疗效果预测的指标。
关于miRNA基因多态性与结直肠癌患者预后 的关系,国内外也有部分学者进行了研究。刑金良 等[24]发现pre-miR-423基因rs6505162位点的多态性 与结直肠癌患者的总生存和无病生存期相关;而pre- miR-608基因rs4919510位点多态性与结直肠癌患者 的无病生存期相关。但这些结果仅在接受化疗的 结直肠癌患者中有意义。在未接受化疗的患者中, 这些多态性均与预后无关。该研究的结果表明, 这些位点有可能通过影响化疗疗效,从而改变患 者的生存预后。在另一个研究中,Lee等[25]发现 miR-492基因rs2289030位点C/G和G/G基因型的结 直肠癌患者,其无病生存期明显短于C/C基因型的 患者,但是其总生存期并无显著差异。以上的研 究结果证明,miRNA基因多态性有可能用于帮助 判断结直肠癌患者的预后。 3 miRNA靶基因多态性与结直肠癌的关系
除了miRNA本身的基因多态性,其靶基因上 与miRNA结合位点的多态性也可能影响结直肠癌 的发病风险。靶基因上miRNA结合位点的多态性 可能削弱miRNA与靶基因的结合或者使得miRNA 与靶基因之间形成一个完美的匹配序列,因而可 能削弱或增强miRNA对靶基因的作用。目前国外 对miRNA结合位点多态性的研究较少。已有的研 究显示,IC53、CD86、INSR等基因的部分多态性 位点与结直肠癌易感性相关[17, 26, 27],部分靶基因结 合位点的多态性在表3中列出。其中,IC53基因的 rs2737位点发生T>C突变时,可形成一个miR-379 的结合位点。miR-379与该位点结合后可抑制IC53 的转录过程,减少了结直肠癌的发病可能;CD86 基因rs17281995位点发生G>C突变时,miR-337、 miR-582和miR-200a与CD86基因的结合削弱,而 miR-184和miR-212与CD86基因的结合增强,最终 导致结直肠癌发病风险升高[17]。
|
|
表3 部分miRNA靶基因多态性与结直肠癌的关系 Table 3 Relation between target gene polymorphism and colorectal cancer |
对于miRNA靶基因多态性与结直肠癌药物治 疗的关系,国外也有少量研究。dhfr基因的rs829 位点为miR-24结合位点,其C>T多态性可能影响 miR-24的功能,从而改变MTX对肿瘤细胞的杀 伤作用[13]。kras基因中一个let-7 miRNA结合位点 (lcs6)则被观察到与kras野生型的转移性结直肠癌 患者使用西妥昔单抗治疗的客观缓解率有关。此 位点T/G或G/G基因型的患者客观缓解率为42%, 而T/T基因型的患者客观缓解率仅为9%。同时, T/G和G/G基因型的患者与T/T基因型的患者相比, 有着更长的中位生存期和总生存期[28]。 4 展望
目前已经明确,miRNA的表达谱改变在结直 肠癌中起着关键性的作用。一系列的研究表明了 不同的miRNA影响着结直肠癌的发生、发展和转移等。最新的研究则提示了miRNA及其靶基因 的多态性也可能影响结直肠癌的发病风险。但目 前在miRNA及其靶基因多态性与结直肠癌的关 系上,国内外的研究仍然较少。希望在不远的将 来,越来越多关于此方面的研究能够不断提升我 们对结直肠癌的认识,为今后的研究打下更为坚 实的基础。
| [1] | Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002[J]. CA Cancer J Clin, 2005,55(2):74-108. |
| [2] | Haggar FA, Boushey RP. Colorectal cancer epidemiology: ncidence, mortality, survival, and risk factors[J]. Clinics Colon ectal Surg, 2009,22(4):191-7. |
| [3] | Dong Y, Wu WK, Wu CW, et al. microRNA dysregulation n colorectal cancer: a clinical perspective[J]. Br J Cancer, 011,104(6):893-8. |
| [4] | Chen L, Wang X, Wang H, et al. miR-137 is frequently down- egulated in glioblastoma and is a negative regulator of COX-2[J]. ur J Cancer, 2012,48(16):3104-11. |
| [5] | Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role n cancer[J]. Nat Rev Cancer, 2006,6(4):259-69. |
| [6] | Zhan JF, Chen LH, Chen ZX, et al. A functional variant n microRNA-196a2 is associated with susceptibility of olorectal cancer in a Chinese population[J]. Arch Med Res, 011,42(2):144-8. |
| [7] | Baraniskin A, Birkenkamp-Demtroder K, Maghnouj A, et al. miR- 0a-5p suppresses tumor growth in colon carcinoma by targeting TL[J]. Carcinogenesis, 2012,33(4):732-9. |
| [8] | Liu K, Li G, Fan C, et al. Increased expression of microRNA-21 nd its association with chemotherapeutic response in human olorectal cancer[J]. J Int Med Res, 2011,39(6):2288-95. |
| [9] | Reid JF, Sokolova V, Zoni E, et al. miRNA profi ling in colorectal ancer highlights miR-1 involvement in met-dependent roliferation[J]. Mol Cancer Res, 2012,10(4):504-15. |
| [10] | Chen ML, Liang LS, Wang XK. miR-200c inhibits invasion and igration in human colon cancer cells sw480/620 by targeting EB1[J]. Clin Exp Metastasis, 2012,29(5):457-69. |
| [11] | Humphreys KJ, Cobiac L, Le Leu RK, et al. Histone deacetylase nhibition in colorectal cancer cells reveals competing roles for embers of the oncogenic miR-17-92 cluster[J]. Mol Carcinog, 013,52(6):459-74. |
| [12] | Li JM, Zhao RH, Li ST, et al. Down-regulation of fecal miR-143 nd miR-145 as potential markers for colorectal cancer[J]. Saudi ed J, 2012,33(1):24-9. |
| [13] | Mishra PJ, Song B, Mishra PJ, et al. miR-24 tumor suppressor ctivity is regulated independent of p53 and through a target site olymorphism[J]. PloS One, 2009,4(12):e8445. |
| [14] | Nagel R, le Sage C, Diosdado B, et al. Regulation of the denomatous polyposis coli gene by the miR-135 family incolorectal cancer[J]. Cancer Res, 2008,68(14):5795-802. |
| [15] | Slaby O, Svoboda M, Fabian P, et al. Altered expression f miR-21, miR-31, miR-143 and miR-145 is related to linicopathologic features of colorectal cancer[J]. Oncology, 007,72(5-6):397-402. |
| [16] | Schetter AJ, Harris CC. Alterations of micrornas contribute to olon carcinogenesis[J]. Semin Oncol, 2011,38(6):734-42. |
| [17] | Landi D, Moreno V, Guino E, et al. Polymorphisms affecting icroRNA regulation and associated with the risk of dietary- elated cancers: a review from the literature and new evidence or a functional role of rs17281995 (cd86) and rs1051690 (insr), reviously associated with colorectal cancer[J]. Mutat Res, 011,717(1-2):109-15. |
| [18] | Kawasaki H, Taira K. microRNA-196 inhibits hoxb8 expression n myeloid differentiation of hl60 cells[J]. Nucleic Acids Symp er (Oxf), 2004,48:211-2. |
| [19] | Min KT, Kim JW, Jeon YJ, et al. Association of the miR-146ac>g, 49c>t, 196a2c>t, and 499a>g polymorphisms with colorectal ancer in the korean population[J]. Mol Carcinog, 2012,51 Suppl :E65-73. |
| [20] | Zhu L, Chu H, Gu D, et al. A functional polymorphism in iRNA-196a2 is associated with colorectal cancer risk in a hinese population[J]. DNA Cell Biol, 2012,31(3):350-4. |
| [21] | Chen H, Sun LY, Chen LL, et al. A variant in microRNA-196a2 is ot associated with susceptibility to and progression of colorectal ancer in Chinese[J]. Intern Med J, 2012,42(6):e115-9. |
| [22] | Guo J, Jin M, Zhang M, et al. A genetic variant in miR-196a2 ncreased digestive system cancer risks: a meta-analysis of 15 ase-control studies[J]. PloS One, 2012,7(1):e30585. |
| [23] | Boni V, Zarate R, Villa JC, et al. Role of primary miRNA olymorphic variants in metastatic colon cancer patients treated ith 5-fluorouracil and irinotecan[J]. Pharmacogenomics J, 011,11(6):429-36. |
| [24] | Xing J, Wan S, Zhou F, et al. Genetic polymorphisms in pre- icroRNA genes as prognostic markers of colorectal cancer[J]. ancer Epidemiol Biomarkers Prev, 2012,21(1):217-27. |
| [25] | Lee HC, Kim JG, Chae YS, et al. Prognostic impact of icroRNA-related gene polymorphisms on survival of atients with colorectal cancer[J]. J Cancer Res Clin Oncol, 010,136(7):1073-8. |
| [26] | Landi D, Gemignani F, Naccarati A, et al. Polymorphisms within icro-RNA-binding sites and risk of sporadic colorectal cancer[J]. arcinogenesis, 2008,29(3):579-84. |
| [27] | Chen J, Shi Y, Li Z, et al. A functional variant of ic53 orrelates with the late onset of colorectal cancer[J]. Mol Med, 011,17(7-8):607-18. |
| [28] | Zhang W, Winder T, Ning Y, et al. A let-7 microRNA-binding ite polymorphism in 3'-untranslated region of kras gene predicts esponse in wild-type kras patients with metastatic colorectal ancer treated with cetuximab monotherapy[J]. Ann Oncol, 011,22(1):104-9. |
| [29] | Akao Y, Nakagawa Y, Naoe T. Let-7 microRNA functions as a otential growth suppressor in human colon cancer cells[J]. Biol harm Bull, 2006,29(5):903-6. |
| [30] | Xi Y, Formentini A, Chien M, et al. Prognostic values of micrornas n colorectal cancer[J]. Biomark Insights, 2006,2:113-21. |
| [31] | Monzo M, Navarro A, Bandres E, et al. Overlapping expression f microRNAs in human embryonic colon and colorectal cancer[J]. Cell Res, 2008,18(8):823-33. |
| [32] | Chang KH, Miller N, Kheirelseid EA, et al. microRNA-21 and DCD4 expression in colorectal cancer[J]. Eur J Surg Oncol, 011,37(7):597-603. |
| [33] | Liu M, Tang Q, Qiu M, et al. miR-21 targets the tumor suppressor hob and regulates proliferation, invasion and apoptosis in olorectal cancer cells[J]. FEBS lett, 2011,585(19):2998-3005. |
| [34] | Yamakuchi M, Yagi S, Ito T, et al. microRNA-22 regulates ypoxia signaling incolon cancer cells[J]. PloS One, 2011,6(5): 20291. |
| [35] | Almeida MI, Nicoloso MS, Zeng L, et al. Strand-specifi c miR-28- p and miR-28-3p have distinct effects in colorectal cancer cells[J]. Gastroenterology, 2012,142(4):886-96,e889. |
| [36] | Kuo TY, Hsi E, Yang IP, et al. Computational analysis of mRNA xpression profiles identifies microRNA-29a/c as predictor f colorectal cancer early recurrence[J]. PloS One, 2012,7(2): 31587. |
| [37] | Yu XF, Zou J, Bao ZJ, et al. miR-93 suppresses proliferation and olony formation of human colon cancer stem cells[J]. World J astroenterol, 2011,17(42):4711-7. |
| [38] | Huang Z, Huang S, Wang Q, et al. MicroRNA-95 promotes cell roliferation and targets sorting Nexin 1 in human colorectal arcinoma[J]. Cancer Res, 2011,71(7):2582-9. |
| [39] | Strillacci A, Griffoni C, Sansone P, et al. MiR-101 downregulation s involved in cyclooxygenase-2 overexpression in human colon ancer cells[J]. Exp Cell Res, 2009,315(8):1439-47. |
| [40] | Wang X, Lam EK, Zhang J, et al. Microrna-122a functions as a ovel tumor suppressor downstream of adenomatous polyposis oli in gastrointestinal cancers[J]. Biochem Biophys Res ommun, 2009,387(2):376-80. |
| [41] | Li XM, Wang AM, Zhang J, et al. Down-regulation of miR-126 xpression in colorectal cancer and its clinical significance[J]. ed Oncol, 2011,28(4):1054-7. |
| [42] | Guo C, Sah JF, Beard L, et al. The noncoding rna, miR-126, uppresses the growth of neoplastic cells by targeting hosphatidylinositol 3-kinase signaling and is frequently ost in colon cancers[J]. Genes Chromosomes Cancer, 008,47(11):939-46. |
| [43] | Hu G, Chen D, Li X, et al. miR-133b regulates the MET proto- ncogene and inhibits the growth of colorectal cancer cells in itro and in vivo[J]. Cancer Biol Ther, 2010,10(2):190-7. |
| [44] | Chen X, Guo X, Zhang H, et al. Role of miR-143 targeting KRAS n colorectal tumorigenesis[J]. Oncogene, 2009,28(10):1385-92. |
| [45] | Ng EK, Tsang WP, Ng SS, et al. MicroRNA-143 targets DNA ethyltransferases 3A in colorectal cancer[J]. Br J Cancer, 009,101(4):699-706. |
| [46] | Akao Y, Nakagawa Y, Naoe T. MicroRNA-143 and -145 in colon ancer[J]. DNA Cell Biol, 2007,26(5):311-20. |
| [47] | Xu N, Papagiannakopoulos T, Pan G, et al. MicroRNA-145 egulates OCT4, SOX2, and KLF4 and represses pluripotency in uman embryonic stem cells[J]. Cell, 2009,137(4):647-58. |
| [48] | Shi B, Sepp-Lorenzino L, Prisco M, et al. Micro RNA 145 targets he insulin receptor substrate-1 and inhibits the growth of colon ancer cells[J]. J Biol Chem, 2007,282(45):32582-90. |
| [49] | Song Y, Xu Y, Wang Z, et al. MicroRNA-148b suppresses cell rowth by targeting cholecystokinin-2 receptor in colorectal ancer[J]. Int J Cancer, 2012,131(5):1042-51. |
| [50] | Karaayvaz M, Pal T, Song B, et al. Prognostic significance f miR-215 in colon cancer[J]. Clin Colorectal Cancer, 011,10(4):340-7. |
| [51] | Liu L, Chen L, Xu Y, et al. microRNA-195 promotes apoptosis nd suppresses tumorigenicity of human colorectal cancer cells[J]. iochem Biophys Res Commun, 2010,400(2):236-40. |
| [52] | Chiang Y, Song Y, Wang Z, et al. Aberrant expression of mir-203 nd its clinical signifi cance in gastric and colorectal cancers[J]. J astrointest Surg, 2011,15(1):63-70. |
| [53] | Sun K, Wang W, Zeng JJ, et al. MicroRNA-221 inhibits DKN1C/p57 expression in human colorectal carcinoma[J]. Acta harmacol Sin, 2011,32(3):375-84. |
| [54] | Zhang Y, He X, Liu Y, et al. microRNA-320a inhibits tumor nvasion by targeting neuropilin 1 and is associated with liver etastasis in colorectal cancer[J]. Oncol Rep, 2012,27(3):685-94. |
| [55] | Wang H, Wu J, Meng X, et al. MicroRNA-342 inhibits colorectal ancer cell proliferation and invasion by directly targeting DNA ethyltransferase 1[J]. Carcinogenesis, 2011,32(7):1033-42. |
| [56] | Nie J, Liu L, Zheng W, et al. microRNA-365, down-regulated n colon cancer, inhibits cell cycle progression and promotes poptosis of colon cancer cells by probably targeting Cyclin D1 nd Bcl-2[J]. Carcinogenesis, 2012,33(1):220-5. |
| [57] | Dai X, Chiang Y, Wang Z, et al. Expression levels of icroRNA-375 in colorectal carcinoma[J]. Mol Med Rep, 012,5(5):1299-304. |
| [58] | Bandres E, Bitarte N, Arias F, et al. Microrna-451 regulates acrophage migration inhibitory factor production and roliferation of gastrointestinal cancer cells[J]. Clin Cancer Res, 009,15(7):2281-90. |
| [59] | Liu X, Zhang Z, Sun L, et al. MicroRNA-499-5p promotes cellular nvasion and tumor metastasis in colorectal cancer by targeting OXO4 and PDCD4[J]. Carcinogenesis, 2011,32(12):1798-805. |
| [60] | Tsang WP, Ng EK, Ng SS, et al. Oncofetal h19-derived mir-675 egulates tumor suppressor rb in human colorectal cancer[J]. arcinogenesis, 2010,31(3):350-8. |
 2014, Vol.41
2014, Vol.41