Correlation of RELL1 Expression with Clinical Pathological Characteristics and Prognosis of Patients with Colon Cancer
-
摘要:目的
探讨RELL1的表达与结肠癌患者临床病理特征以及预后的关系。
方法选择80例结肠癌组织及80例癌旁组织的组织芯片进行RELL1蛋白的免疫组织化学实验。比较RELL1蛋白不同表达水平与不同临床病理参数之间的关系,单因素和多因素Cox风险比例回归分析结肠癌患者生存的影响因素,Kaplan-Meier生存曲线分析RELL1表达与结肠癌患者生存率的关系,Log rank检验生存率差异。
结果结肠癌组织中RELL1表达低于癌旁组织(P<0.05)。癌组织中RELL1的表达与TNM分期、N分期有一定相关性(P<0.05)。 RELL1高表达的结肠癌患者3年总生存率(OS)高于RELL1低表达的结肠癌患者(P<0.05)。多因素Cox回归分析显示,RELL1低表达、高龄及高TNM分期是结肠癌患者预后不良的危险因素(P<0.05)。
结论结肠癌组织中RELL1表达下调, RELL1低表达、高龄和高TNM期可导致患者不良结局。
Abstract:ObjectiveTo explore the correlation of RELL1 expression with clinical pathological features and prognosis of patients with colon cancer.
MethodsImmunohistochemical experiments of the RELL1 protein were performed on tissue chips from 80 colon cancer tissues and 80 adjacent tissues. The relationship between different expression levels of RELL1 protein and clinical pathological parameters was compared. Univariate and multivariate Cox risk proportional regression analyses were conducted on factors affecting the survival of patients with colon cancer. Kaplan-Meier survival curve analysis was conducted on the survival rates of patients with colon cancer and different levels of RELL1 expression. Log rank test was performed to evaluate differences in survival rates.
ResultsThe expression of RELL1 in colon cancer tissues was lower than that in adjacent tissues (P<0.05). The expression of RELL1 in cancer tissues is correlated with TNM stage and N stage (P<0.05). The 3-year overall survival (OS) rate of colon cancer patients with high RELL1 expression was higher than that of patients with low RELL1 expression (P<0.05). Multivariate Cox regression analysis showed that low RELL1 expression, advanced age, and high TNM stage were risk factors for poor prognosis in patients with colon cancer (P<0.05).
ConclusionThe expression of RELL1 is downregulated in colon cancer tissue, and the low RELL1 expression, advanced age, and high TNM stage can lead to adverse outcomes in patients.
-
Key words:
- Colon cancer /
- RELL1 /
- Clinical pathological features /
- Prognosis
-
0 引言
结直肠癌(Colorectal cancer,CRC)是全球最常见的肿瘤之一,2018年全球癌症相关死亡病因中居第二位[1],在我国是仅次于肺癌、胃癌的第三大高发癌症[2]。多数结肠癌患者会出现肿瘤复发,预后较差[3]。淋巴组织表达受体(Receptor expressed in lymphoid tissues, RELT)是一种肿瘤坏死因子家族成员,与其两个类似的RELL1和RELL2统称为RELTfms[4]。近年RELTfms与癌症的联系引起了较多关注。RELL1为蛋白质编码基因[5],其蛋白结构特点是包含典型的胞外半胱氨酸残基,但不包含一些TNF受体家族成员中存在的保守的胞内死亡结构[6]。TNF受体家族成员可以诱导应激反应、炎性反应、迁移和增殖等细胞过程[7]。RELL1通过与TNF受体相关因子1(TNF receptor-associated factor 1, TRAF1)结合来激活促炎通路[8]。也有报道RELT家族成员通过不同TNF受体的机制激活p38并诱导细胞凋亡[9]。本研究前期发现RELL1的表达水平在结肠腺瘤进展至结肠癌过程中有逐渐下降的趋势,且RELL1与结肠肿瘤之间报道较少,故进一步探讨RELL1的表达与结肠癌患者临床病理特征及预后的关系,旨在为临床诊治和预后评估提供参考。
1 资料与方法
1.1 生物信息学方法分析RELL1表达量
从TCGA数据库(https://portal.gdc.cancer.gov)下载并整理结肠癌的RNAseq数据,从UCSC XENA(https://xenabrowser.net/datapages)提取TCGA中的结肠癌数据以及GTEx中对应的正常组织数据,从GEO数据库(https://www.ncbinlm.nih.gov/geo)下载GSE20916的基因表达谱,提取并分析RELL1基因在结肠癌组织与正常组织中的表达差异。
1.2 生物信息学方法探究RELL1在结肠癌中的生存分析
利用在线GEPIA数据库(http://gepia.cancer-pku.cn)、HPA数据库(https://www.proteinatlas.org)选择结肠肿瘤、RELL1基因进行生存分析曲线绘制。下载GSE17536数据提取生存时间及状态,使用Cox回归方法对结肠肿瘤中RELL1高低分组绘制生存曲线,探讨RELL1在结肠癌中的意义。
1.3 组织样本来源
结肠癌组织芯片(购自武汉大械生物医疗科技有限公司)共包括80例结肠癌样本及80例癌旁组织,均有完整的病例信息,包括性别、年龄、病理分期、生存及随访信息。OS时间定义为自确诊至死亡或随访截止时间。本研究已通过医院伦理审批免知情同意(2023-078)。
1.4 免疫组织化学实验及半定量分析
RELL1抗体购自英国Abcam公司,采用EnVisionTM两步法批量检测组织芯片RELL1蛋白表达。微波加热抗原修复,3%H2O2封闭内源性过氧化物酶,滴加一抗(1∶100),4℃冰箱过夜,PBS缓冲液清洗,滴加二抗 (1∶100),DAB显色,苏木精对比染色,常规封片。采用免疫反应性评分方法(Immunoreactive score, IRS)作为评分标准,显微镜下随机选择不重叠的3个视野,人工计数阳性着色细胞,在光学显微镜下对组织切片分别按染色程度(阴性着色、淡黄色、浅褐色、深褐色分别为0、1、2、3分)、阳性范围进行评分(1~4分别为0~25%、26%~50%、51%~75%、76%~100%),最后进行染色程度与阳性范围评分相乘得到IRS范围(0~12分)。由两名资深病理学医师在双盲条件下独立观察染色结果。当存在争议时,引入上级医院作为第三方判断。
1.5 统计学方法
使用SPSS软件(22.0版和R 4.2.2版)进行统计分析。正态分布的计量资料用均数±标准差表示,非正态分布的计量资料用中位数表示。Student-t检验和皮尔逊卡方检验用于比较组间差异。绘制Kaplan-Meier曲线比较组间预后差异,Log rank检验生存率差异。通过单因素和多因素Cox比例风险回归分析影响结肠癌患者生存的因素,其中将单因素分析结果中P<0.1的变量纳入多因素分析。基于多因素Cox回归模型的结果,使用Nomogram绘制列线图,采用Bootstrap自抽样法进行内部验证,计算一致性指数,绘制校准曲线并对列线图进行验证,P<0.05为差异有统计学意义。
2 结果
2.1 RELL1在不同数据库中的表达结果及RELL1在结肠癌中的生存分析
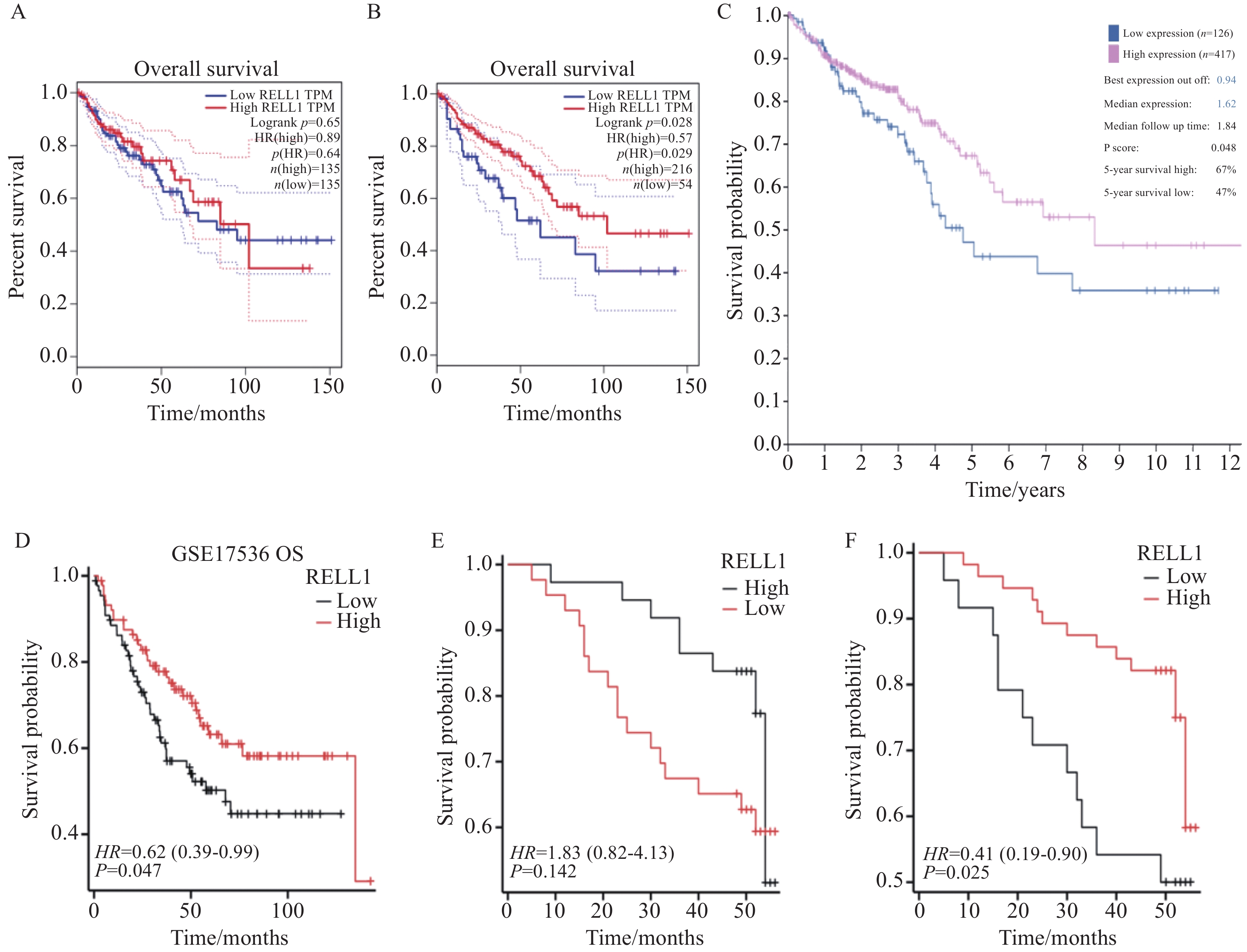
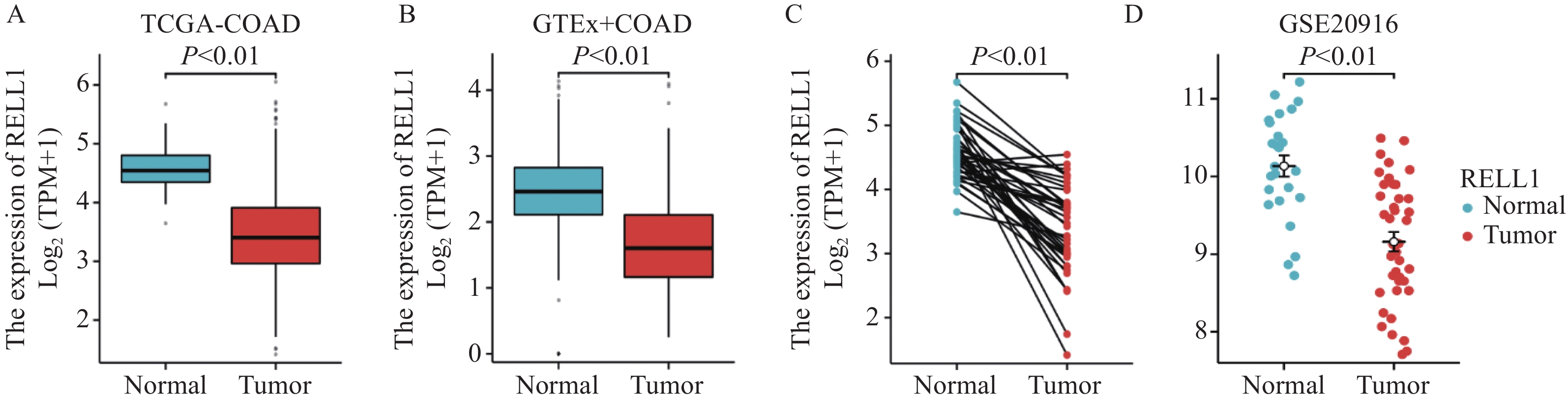
多个数据库分析发现RELL1在结肠癌中的表达较正常组明显下降,差异有统计学意义(P<0.001),见图1。在GEPIA数据库中,当Cut off值为中位数时,KM曲线中生存率无统计学差异,见图2A,当进一步选择下调RELL1表达值,把Cut off值调为20%时,我们发现生存率差异有统计学意义(P<0.05),见图2B。同时在HPA数据库中也发现当RELL1 mRNA取值在0.94时(低表达例数∶高表达例数为126∶471),生存率差异有统计学意义(P<0.05),见图2C。当然,这两个数据库均是基于TCGA数据库统计,我们同时也探索GEO数据库中GSE17536数据集,包括结肠癌样本177例,按照RELL1表达量中位数作为高低分组即发现高表达组OS生存率高于低表达组,见图2D。
![]() 图 2 RELL1高低表达的结肠肿瘤患者的OS生存曲线Figure 2 OS survival curves of patients with colon cancer based on high or low expression of RELL1A: RELL1 OS survival curve in the GEPIA database (50% cut off); B: RELL1 OS survival curve in the GEPIA database (20% cut off); C: RELL1 OS survival curve in the HPA database (cut off value 0.94); D: RELL1 OS survival curve in GSE17536 (50% cut off); E: survival curve after median IRS grouping of colon cancer tissue chips (IRS=6); F: survival curve after optimal IRS grouping of colon cancer tissue chips (IRS=4).
图 2 RELL1高低表达的结肠肿瘤患者的OS生存曲线Figure 2 OS survival curves of patients with colon cancer based on high or low expression of RELL1A: RELL1 OS survival curve in the GEPIA database (50% cut off); B: RELL1 OS survival curve in the GEPIA database (20% cut off); C: RELL1 OS survival curve in the HPA database (cut off value 0.94); D: RELL1 OS survival curve in GSE17536 (50% cut off); E: survival curve after median IRS grouping of colon cancer tissue chips (IRS=6); F: survival curve after optimal IRS grouping of colon cancer tissue chips (IRS=4).2.2 病例资料
纳入80例结肠癌样本,中位随访48(5~56)个月。患者诊断时的中位年龄为61岁,其中35例(43.8%)患者年龄≥60岁。男性43例(53.8%),女性37例(46.2%)。TNM分期Ⅰ~Ⅱ、Ⅲ~Ⅳ期肿瘤患者分别为46例(57.5%)、34例(42.5%)。80例CRC患者的详细临床参数见表1。
表 1 RELL1表达与结肠癌患者临床病理参数的关系Table 1 Relationship between RELL1 expression and clinical pathological parameters of colon cancer patientsCharacteristics High RELL1
expression (n=56)Low RELL1
expression (n=24)P Gender 0.129 Female 29(36.2%) 8(10.0%) Male 27(33.8%) 16(20.0%) Age(years) 0.806 ≤60 31(38.8%) 14(17.5%) >60 25(31.2%) 10(12.5%) T stage 0.151 T1-2 11(13.8%) 1(1.2%) T3-4 45(56.2%) 23(28.7%) N stage 0.042 N0 37(46.2%) 10(12.5%) N1-2 19(23.8%) 14(17.5%) M stage 0.539 M0 49(61.3%) 19(23.8%) M1 7(8.8%) 5(6.2%) TNM stage 0.018 Ⅰ-Ⅱ 37(46.2%) 9(11.2%) Ⅲ-Ⅳ 19(23.8%) 15(18.8%) Tumor size(cm) 0.304 >5 28(35.0%) 15(18.8%) ≤5 28(35.0%) 9(11.2%) Differentiation 0.512 Well and moderate 43(53.8%) 20(25.0%) Poor 13(16.2%) 4(5.0%) 2.3 RELL1表达与临床病理参数的关系以及对结肠癌患者生存分析的影响
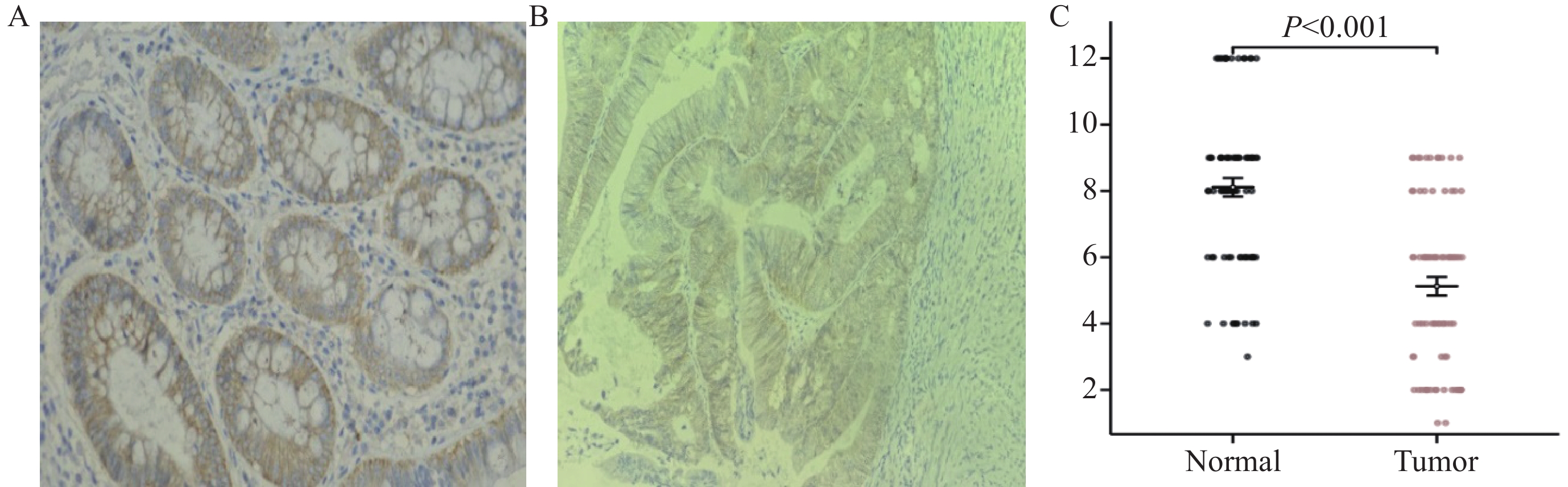
免疫组织化学实验结果显示, RELL1在癌组织和癌旁组织中的IRS平均值分别为5.13±2.51和8.11±2.50分(P<0.001),见图3 。
![]() 图 3 RELL1蛋白在结肠癌组织和癌旁组织中的表达(IHC ×400)Figure 3 Expression of RELL1 protein in colon cancer tissues and adjacent tissues (IHC ×400)A: high expression of the RELL1 protein in normal colon tissue; B: low expression of the RELL1 protein in colon cancer tissue; C: IRS expression in colon cancer tissue is significantly lower than that in adjacent tissue.
图 3 RELL1蛋白在结肠癌组织和癌旁组织中的表达(IHC ×400)Figure 3 Expression of RELL1 protein in colon cancer tissues and adjacent tissues (IHC ×400)A: high expression of the RELL1 protein in normal colon tissue; B: low expression of the RELL1 protein in colon cancer tissue; C: IRS expression in colon cancer tissue is significantly lower than that in adjacent tissue.80例CRC组织RELL1的IRS评分中位数为6分,以此作为RELL1表达高低分组,发现生存率差异无统计学意义,见图2E。使用R语言survival包的surv_cutpoint函数测得最佳Cut off值为4分,取IRS≥4表示RELL1高表达(n=56),IRS<4表示RELL1低表达(n=24)时,发现两组生存率差异有统计学意义,见图2F。RELL1蛋白差异表达与患者N分期及TNM分期有相关性(P<0.05),而与患者年龄、性别、T分期、M分期无显著相关性(P>0.05),见表1。
2.4 结肠癌患者预后影响因素分析
以结肠癌患者预后为因变量 (0=存活,1=死亡),以RELL1表达IRS值、年龄、性别、肿瘤直径、分化程度、T分期、N分期、M分期、TNM分期为自变量,单因素Cox回归分析显示高N分期、高TNM分期, 低RELL1表达、高龄与结肠癌患者预后不良有关(P<0.1),多因素Cox回归分析显示高龄、高TNM分期、低RELL1表达是结肠癌患者预后不良的危险因素 (P<0.05),见表2。
表 2 影响结肠癌患者预后的单因素及多因素Cox回归分析Table 2 Univariate and multivariate Cox regression analyses of factors affecting the prognosis of patients with colon cancerCharacteristics n Univariate analysis Multivariate analysis HR(95%CI) P HR(95%CI) P RELL1 expression <0.001 <0.001 High 56 Reference Reference Low 24 4.080(2.002-8.316) 3.915(1.858-8.251) Gender 0.991 Female 37 Reference Male 43 1.004(0.493-2.045) Age(years) 0.089 0.031 ≤60 45 Reference Reference >60 35 1.861(0.910-3.804) 2.288(1.081-4.841) T stage 0.360 T1-2 12 Reference T3-4 68 1.746(0.529-5.762) N stage 0.009 0.177 N0 47 Reference Reference N1-2 33 2.627(1.279-5.392) 0.479(0.164-1.395) M stage 0.266 M0 68 Reference M1 12 1.662(0.679-4.067) TNM stage <0.001 <0.001 Ⅰ-Ⅱ 46 Reference Reference Ⅲ-Ⅳ 34 4.626(2.156-9.924) 6.867(2.221-21.228) Tumor size(cm) 0.135 >5 43 Reference ≤5 37 0.570(0.273-1.192) Differentiation 0.689 Well and moderate 63 Reference Poor 17 0.834(0.342-2.034) 2.5 列线图绘制及验证
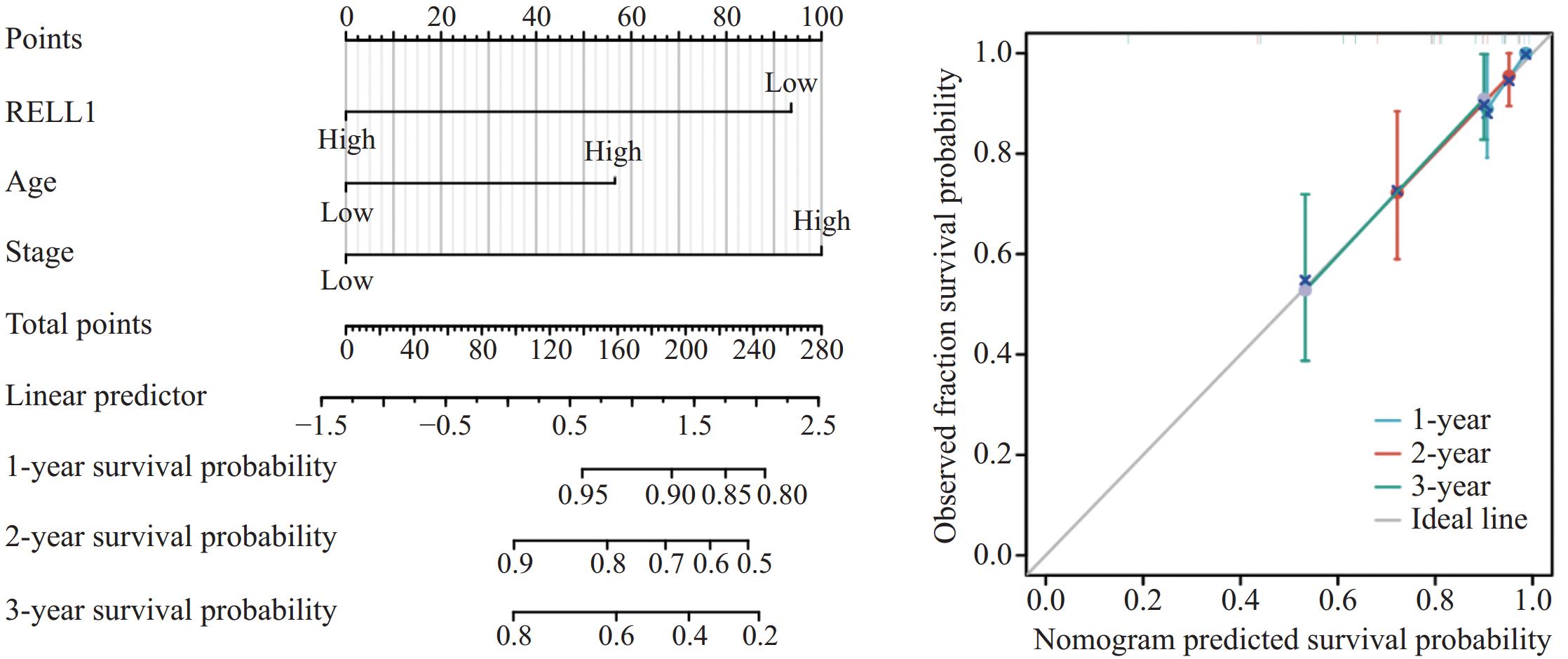
单因素和多因素Cox回归分析均显示RELL1表达、年龄、TNM分期是影响预后的因素,基于这3个因素绘制列线图,用于预测3年内的生存率。校准曲线提示预测风险接近实际风险(C-index:0.788),提示RELL1是预测CRC生存分析的可靠生物标志物,见图4。
3 讨论
结直肠癌的病因及其发生发展机制目前仍不是很清楚,缺乏有效的预防及筛查方法以及特异性诊断和预后指标。我们前期通过生信方法在结肠腺瘤中筛选到RELL1,并进一步研究其在结肠癌中的作用。既往的报道中,RELTfms在不同肿瘤中发挥促癌作用还是抑癌作用存在显著差异。在胶质瘤中RELL1表达上调,并与严重的胶质瘤病理和免疫抑制环境相关,可能是胶质瘤的预后标志物[10-11]。RELT促进恶性B细胞淋巴瘤进展,与健康淋巴结相比,B细胞淋巴瘤活检组织中RELT蛋白表达上调[6]。在非急性早幼粒细胞白血病中,RELL2与不良预后相关[12]。在乳腺癌中RELT与人三阴性乳腺癌中上调的基因簇相关,促进乳腺癌的进展[13]。RELT在小细胞肺癌低危组表达较高,提示了对肺癌的保护作用[14] ,但RELT又是肾癌中的不良预后指标[15]。RELTfms在消化道肿瘤中发挥的作用差异也较大,RELL1在胃肿瘤中发挥抑制胃癌进展的作用,它被转录成一种环状RNA,与胃癌预后不良相关[16]。RELT通过激活NF-kappaB通路促进食管鳞状细胞癌的生长[17],然而RELL2在致瘤性ESCC细胞系中表达水平较低,对食管癌又具有保护作用 [18]。RELL2在胰腺导管癌中也表现出了抗肿瘤作用[19]。有文献通过生物信息学分析表明[20],RELL1可能通过竞争miRNA导致结直肠癌基因表达失调从而促进肿瘤进展,这与我们的结果相反,但其结论缺乏实验验证,具体机制有待进一步探讨。
我们通过挖掘多个数据库联合分析发现,与正常组相比RELL1 mRNA的表达在结肠癌中明显下降,推测其有可能起到一定抑癌作用,故进一步探索RELL1在结肠癌中对生存预后的影响。在GEPIA及HPA数据库中发现按照较低的RELL1水平进行高低分组对结肠癌患者的生存率影响有统计学意义,并通过组织芯片免疫组织化学实验进一步验证。在研究中发现癌旁组织RELL1蛋白的IRS分数明显高于结肠癌组织。按照RELL1的IRS评分中位数进行RELL1表达高低分组,发现生存率虽然无统计学差异,但RELL1高表达患者整体生存趋势较好,而以IRS 4分调整高低分组例数再次分组后,生存率则达到了统计学差异,提示RELL1蛋白的表达对结肠癌生存率有一定影响。RELL1蛋白在高TNM分期及高N分期肿瘤中的低表达比例更高(P<0.05),提示随着结肠肿瘤分期的进展,RELL1表达有下降趋势。本研究结果推测RELL1低表达到一定程度后,可能通过某种方式如免疫抑制或者肿瘤信号通路的激活促进结肠癌的进展。通过单因素及多因素Cox回归分析发现结肠癌组织中低RELL1、高TNM分期、高龄是结肠癌预后不良的危险因素,并绘制预后列线图进一步预测了患者1、2及3年的生存预测。
综上,本研究运用生信分析及组织芯片实验相结合,初步探索了RELL1在结肠癌中的表达及对患者预后的影响。但仍存在一些局限性,比如样本数的限制、实验方法较单一,尚不能直接证实 RELL1在结肠癌中的致病机制,其中涉及的信号通路等分子机制仍有待进一步研究,未来仍需开展大样本临床研究和细胞及动物等基础研究证实。
Competing interests: The authors declare that they have no competing interests.利益冲突声明:所有作者均声明不存在利益冲突。作者贡献:冯 洁:课题设计,文章撰写与修改张亚男:实验实施程诺、楚艳、沈云海:数据分析统计,文献调研潘勤聪:课题指导,文章修改指导 -
表 1 RELL1表达与结肠癌患者临床病理参数的关系
Table 1 Relationship between RELL1 expression and clinical pathological parameters of colon cancer patients
Characteristics High RELL1
expression (n=56)Low RELL1
expression (n=24)P Gender 0.129 Female 29(36.2%) 8(10.0%) Male 27(33.8%) 16(20.0%) Age(years) 0.806 ≤60 31(38.8%) 14(17.5%) >60 25(31.2%) 10(12.5%) T stage 0.151 T1-2 11(13.8%) 1(1.2%) T3-4 45(56.2%) 23(28.7%) N stage 0.042 N0 37(46.2%) 10(12.5%) N1-2 19(23.8%) 14(17.5%) M stage 0.539 M0 49(61.3%) 19(23.8%) M1 7(8.8%) 5(6.2%) TNM stage 0.018 Ⅰ-Ⅱ 37(46.2%) 9(11.2%) Ⅲ-Ⅳ 19(23.8%) 15(18.8%) Tumor size(cm) 0.304 >5 28(35.0%) 15(18.8%) ≤5 28(35.0%) 9(11.2%) Differentiation 0.512 Well and moderate 43(53.8%) 20(25.0%) Poor 13(16.2%) 4(5.0%) 表 2 影响结肠癌患者预后的单因素及多因素Cox回归分析
Table 2 Univariate and multivariate Cox regression analyses of factors affecting the prognosis of patients with colon cancer
Characteristics n Univariate analysis Multivariate analysis HR(95%CI) P HR(95%CI) P RELL1 expression <0.001 <0.001 High 56 Reference Reference Low 24 4.080(2.002-8.316) 3.915(1.858-8.251) Gender 0.991 Female 37 Reference Male 43 1.004(0.493-2.045) Age(years) 0.089 0.031 ≤60 45 Reference Reference >60 35 1.861(0.910-3.804) 2.288(1.081-4.841) T stage 0.360 T1-2 12 Reference T3-4 68 1.746(0.529-5.762) N stage 0.009 0.177 N0 47 Reference Reference N1-2 33 2.627(1.279-5.392) 0.479(0.164-1.395) M stage 0.266 M0 68 Reference M1 12 1.662(0.679-4.067) TNM stage <0.001 <0.001 Ⅰ-Ⅱ 46 Reference Reference Ⅲ-Ⅳ 34 4.626(2.156-9.924) 6.867(2.221-21.228) Tumor size(cm) 0.135 >5 43 Reference ≤5 37 0.570(0.273-1.192) Differentiation 0.689 Well and moderate 63 Reference Poor 17 0.834(0.342-2.034) -
[1] Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2018, 68(6): 394-424. doi: 10.3322/caac.21492
[2] 中国抗癌协会大肠癌专业委员会中国结直肠肿瘤早诊筛查策略制订专家组. 中国结直肠肿瘤早诊筛查策略专家共识[J]. 中华胃肠外科杂志, 2018, 21(10): 1081-1086. [Expert Group of Early Diagnosis and Screening Strategies Development for Colorectal Tumors in China in the Colorectal Cancer Professional Committee of the Chinese Anti Cancer Association. Expert Consensus on Early Diagnosis and Screening Strategies for Colorectal Tumors in China[J]. Zhonghua Wei Chang Wai Ke Za Zhi, 2018, 21(10): 1081-1086.] Expert Group of Early Diagnosis and Screening Strategies Development for Colorectal Tumors in China in the Colorectal Cancer Professional Committee of the Chinese Anti Cancer Association. Expert Consensus on Early Diagnosis and Screening Strategies for Colorectal Tumors in China[J]. Zhonghua Wei Chang Wai Ke Za Zhi, 2018, 21(10): 1081-1086.
[3] Benson AB, Venook AP, Al-Hawary MM, et al. NCCN Guidelines Insights: Colon Cancer, Version 2. 2018[J]. J Natl Compr Canc Netw, 2018, 16(4): 359-369.
[4] Cusick JK, Alcaide J, Shi Y, et al. The RELT Family of Proteins: An Increasing Awareness of Their Importance for Cancer, the Immune System, and Development[J]. Biomedicines, 2023, 11(10): 2695. doi: 10.3390/biomedicines11102695
[5] Cusick JK, Xu LG, Bin LH, et al. Identification of RELT homologues that associate with RELT and are phosphorylated by OSR1[J]. Biochem Biophys Res Commun, 2006, 340(2): 535-543. doi: 10.1016/j.bbrc.2005.12.033
[6] Cusick JK, Alhomsy Y, Wong S, et al. RELT stains prominently in B-cell lymphomas and binds the hematopoietic transcription factor MDFIC[J]. Biochem Biophys Rep, 2020, 24: 100868.
[7] Dostert C, Grusdat M, Letellier E, et al. The TNF Family of Ligands and Receptors: Communication Modules in the Immune System and Beyond[J]. Physiol Rev, 2019, 99(1): 115-160. doi: 10.1152/physrev.00045.2017
[8] Sica GL, Zhu G, Tamada K, et al. RELT, a new member of the tumor necrosis factor receptor superfamily, is selectively expressed in hematopoietic tissues and activates transcription factor NF-kappaB[J]. Blood, 2001, 97(9): 2702-2707. doi: 10.1182/blood.V97.9.2702
[9] Moua P, Checketts M, Xu LG, et al. RELT family members activate p38 and induce apoptosis by a mechanism distinct from TNFR1[J]. Biochem Biophys Res Commun, 2017, 491(1): 25-32. doi: 10.1016/j.bbrc.2017.07.022
[10] Jin X, Xie H, Liu X, et al. RELL1, a novel oncogene, accelerates tumor progression and regulates immune infiltrates in glioma[J]. Int Immunopharmacol, 2020, 87: 106707. doi: 10.1016/j.intimp.2020.106707
[11] Rose M, Cardon T, Aboulouard S, et al. Surfaceome Proteomic of Glioblastoma Revealed Potential Targets for Immunotherapy[J]. Front Immunol, 2021, 12: 746168. doi: 10.3389/fimmu.2021.746168
[12] Zhao Y, Niu LT, Hu LJ, et al. Comprehensive analysis of ECHDC3 as a potential biomarker and therapeutic target for acute myeloid leukemia: Bioinformatic analysis and experimental verification[J]. Front Oncol, 2022, 12: 947492. doi: 10.3389/fonc.2022.947492
[13] Johansson J, Tabor V, Wikell A, et al. TGF-beta1-Induced Epithelial-Mesenchymal Transition Promotes Monocyte/Macrophage Properties in Breast Cancer Cells[J]. Front Oncol, 2015, 5: 3.
[14] Zhang Z, Wu P, Zhang C, et al. Tumor Necrosis Factor Family Member Profile Predicts Prognosis and Adjuvant Chemotherapy Benefit for Patients with Small-Cell Lung Cancer[J]. Front Immunol, 2021, 12: 745769.
[15] Cui Y, Shen T, Xu F, et al. KCNN4 may weaken anti-tumor immune response via raising Tregs and diminishing resting mast cells in clear cell renal cell carcinoma[J]. Cancer Cell Int, 2022, 22(1): 211. doi: 10.1186/s12935-022-02626-7
[16] Sang H, Zhang W, Peng L, et al. Exosomal circRELL1 serves as a miR-637 sponge to modulate gastric cancer progression via regulating autophagy activation[J]. Cell Death Dis, 2022, 13(1): 56. doi: 10.1038/s41419-021-04364-6
[17] Yao W, Chen Q, Li S, et al. RELT promotes the growth of esophageal squamous cell carcinoma by activating the NF-kappaB pathway[J]. Cell Cycle, 2021, 20(13): 1231-1241. doi: 10.1080/15384101.2021.1924451
[18] Tang SJ, Shen H, An O, et al. Cis-and trans-regulations of pre-mRNA splicing by RNA editing enzymes influence cancer development[J]. Nat Commun, 2020, 11(1): 799. doi: 10.1038/s41467-020-14621-5
[19] Li Z, Qin C, Zhao B, et al. DHX38 restricts chemoresistance by regulating the alternative pre-mRNA splicing of RELL2 in pancreatic ductal adenocarcinoma[J]. PLoS Genet, 2023, 19(7): e1010847. doi: 10.1371/journal.pgen.1010847
[20] Kadkhoda S, Darbeheshti F, Rezaei N, et al. Investigation of circRNA-miRNA-mRNA network in colorectal cancer using an integrative bioinformatics approach[J]. Gastroenterol Hepatol Bed Bench, 2021, 14(2): 141-153.



 下载:
下载: