Mendelian Randomized Study of Protective Effect of Statins on Breast Cancer
-
摘要:目的
旨在从遗传学角度探讨他汀类药物对乳腺癌的保护作用。
方法从既往的表达数量性状基因座(eQTL)研究中提取他汀类药物靶基因HMGCR及其他五种胆固醇调控基因(LDLR、PCSK9、ABCG8、APOB及NPC1L1)的工具变量;以这些工具变量预测的胆固醇调控基因作为暴露因素,采用基于汇总数据的孟德尔随机化法(SMR)探讨暴露因素对全部乳腺癌、雌激素受体阳性(ER+)乳腺癌及ER−乳腺癌发病风险的遗传影响。从既往人类全基因组关联研究(GWAS)中提取总胆固醇(TC)、低密度脂蛋白胆固醇(LDL-C)及非高密度脂蛋白胆固醇(non-HDL-C)的工具变量,限定这些变量在染色体上位于上述胆固醇调控基因周围100 kb范围内,从而使这些工具变量可预测上述胆固醇相关基因调控下的TC、LDL-C或non-HDL-C水平,并以此作为暴露因素;采用两样本孟德尔随机化法(IVW、MR-PRESSO及MR-Egger)探讨暴露因素对全部乳腺癌、ER+乳腺癌及ER−乳腺癌发病风险的遗传影响。
结果SMR分析报告显示,HMGCR表达升高与全部乳腺癌及ER+乳腺癌的发病风险增加显著相关(P=0.044,P=0.039),但与ER−乳腺癌的发病风险变化无显著相关性(P=0.190);其他五种调控基因与全部乳腺癌、ER+乳腺癌及ER−乳腺癌的发病风险变化均无显著相关性(均P>0.05)。IVW分析报告显示,在HMGCR的调控下,外周血TC、LDL-C及nonHDL-C水平升高显著增加全部乳腺癌发病风险(P=1.160e-05,P=1.248e-05,P=1.869e-05),亦显著增加ER+乳腺癌发病风险(P=3.181e-04,P=2.231e-04,P=3.520e-04),但与ER−乳腺癌的发病风险变化无关(P=0.062,P=0.133,P=0.055)。MR-PRESSO和MR-Egger分析结果支持IVW结果。
结论他汀类药物的使用可在基因水平上降低ER+乳腺癌的发病风险,而对ER−乳腺癌无此类保护作用。
Abstract:ObjectiveTo genetically investigate the protective effects of statins on breast cancer.
MethodsInstrumental variables for the statin target gene HMGCR and five other cholesterol-regulated genes (LDLR, PCSK9, ABCG8, APOB, and NPC1L1) were obtained from previous expression quantitative trait locus (eQTL) studies. Cholesterol-regulated genes predicted by these instrumental variables served as the exposure factors. Mendelian randomization based on pooled data (SMR) was conducted to explore the genetic effects of exposure factors on the incidence risk of all breast cancers, ER+ breast cancer, and ER-breast cancer. Instrumental variables for total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), and non-high-density lipoprotein cholesterol (non-HDL-C) were derived from a previous human genome-wide association study and restricted to be chromosomally located within 100 kb of the above cholesterol regulatory genes; the instrumental variables could predict TC, LDL-C, or non-HDL-C levels under the regulation of the abovementioned cholesterol-associated genes which were used as exposure factors. Two-sample Mendelian randomization (IVW, MR-PRESSO, and MR-Egger) was used to explore the genetic effects of exposure factors on the risk of all breast cancers, ER+ breast cancer, and ER− breast cancer.
ResultsSMR analysis reported that elevated HMGCR expression was significantly associated with the increased incidence risk of all breast cancers and ER+ breast cancer (P=0.044 and P=0.039, respectively) but not with the change in incidence risk of ER− breast cancer (P=0.190); the other five regulatory genes were not significantly correlated with the change in incidence risk of all breast cancers, ER+ breast cancer, and ER− breast cancer (all P>0.05). IVW analysis reported that under the regulation of HMGCR, elevated levels of peripheral TC, LDL-C, and non-HDL-C significantly increased the incidence risk of all breast cancers (P=1.160e-05, P=1.248e-05, and P=1.869e-05) and the incidence risk of ER+ breast cancer (P=3.181e-04, P=2.231e-04, and P=3.520e-04), but they were not associated with a change in the incidence risk of ER− breast cancer (P=0.062, P=0.133, and P=0.055). The results of MR-PRESSO and MR-Egger analyses supported the IVW results.
ConclusionStatins could reduce the incidence risk of ER+ breast cancer at the genetic level, but there is no such protective effects on ER− breast cancer.
-
Key words:
- Breast cancer /
- Statins /
- HMGCR /
- Mendelian randomization
-
0 引言
在全球女性人群中,乳腺癌是最常见恶性肿瘤类型,也是癌症致死的主要原因之一。最新统计数据显示,全球每年新增乳腺癌病例逾230万,死亡病例逾66万[1]。由于经济发展不平衡、生活方式等因素的持续影响,预计未来全球乳腺癌发病率将继续上升。据估计,到2040年全球每年新发乳腺癌病例将增至约300万,死亡病例数约达100万[2]。近年来,中国乳腺癌发病率增长明显快于全球多数国家。2020年中国新发乳腺癌病例达40万,死亡病例逾10万[3]。若不能有效防控,中国乳腺癌新发和死亡病例将进一步增加。因此,乳腺癌是亟待解决的全球性健康挑战。
学界一直致力于研究乳腺癌发病机制,并不断创新治疗方法[4]。近年来,胆固醇代谢在乳腺癌发生和发展中的作用及其治疗意义被广泛关注。既往研究显示,一些主要类型的胆固醇分子,如低密度脂蛋白胆固醇(Low-density lipoprotein cholesterol, LDL-C)可能与乳腺癌发病风险增加有关[5-6]。这些病理生理机制可能包括参与雌激素合成、促进炎性反应和氧化应激过程以及促进某些肿瘤相关因子的生成[7]。他汀类药物是目前最常用的降胆固醇药物,其通过竞争性抑制内源性胆固醇合成限速酶(HMG-CoA还原酶)来阻断细胞内胆固醇合成[8]。此外,其还有抗炎、抗氧化应激等多种生物学作用,并在恶性肿瘤防治方面具有潜在价值[8]。一项荟萃分析显示,他汀类药物可改善乳腺癌患者预后,包括降低复发风险、全因死亡率和癌症特异性死亡率,但其对乳腺癌发病风险的潜在影响仍需进一步研究[9]。
本研究采用药靶孟德尔随机化经典方法,在基因水平上探讨他汀类药物对乳腺癌的保护作用。除研究他汀类药物的靶基因HMG-CoA外,还扩展至其他五种胆固醇代谢相关调控基因(LDLR、PCSK9、ABCG8、APOB及NPC1L1)对乳腺癌发病风险的影响。而医学界已经或有望基于这些基因研发新一代降胆固醇药物[10]。鉴于雌激素受体(Estrogen receptor, ER)在乳腺癌发病机制、疾病分类及治疗选择中的重要地位,本研究将乳腺癌分为全部乳腺癌、ER+乳腺癌及ER−乳腺癌分别进行研究[11]。这将有助于深入理解胆固醇代谢在乳腺癌发生中的机制,并可能为新的防治策略提供理论依据。
1 资料与方法
1.1 数据汇总
胆固醇调控基因HMGCR和LDLR的汇总数据来自eQTLGen开展的一项大规模顺式和反式表达数量性状基因座(eQTL)分析。该分析包含31 684名欧洲受试者(不限性别),确定数千个调控血液基因表达的基因位点[12]。胆固醇调控基因PCSK9、ABCG8、APOB及NPC1L1的汇总数据则来源于GTEx开展的大规模eQTL分析。GTEx分析涵盖838名欧洲遗体捐献者(不限性别)的49种组织的15 201份样本,全面描述基因对人体组织转录组的影响,并将这些调控机制与性状/疾病联系起来[13]。
三种胆固醇表型:总胆固醇(Total cholesterol, TC)、LDL-C及非高密度脂蛋白胆固醇(Non-high-density lipoprotein cholesterol, nonHDL-C)的汇总数据来自全球血脂遗传学联合会开展的一项人类全基因组关联研究(GWAS)。该研究包含1 320 016名欧洲受试者(不限性别),探讨血脂遗传多样性的价值[14]。全部乳腺癌、ER+乳腺癌及ER−乳腺癌的汇总数据来源于乳腺癌协会联合会开展的GWAS研究。该研究包含来自欧洲人群的122 977名乳腺癌病例和105 974名健康对照受试者(不限性别),确定65个新的乳腺癌风险基因位点[15]。上述汇总数据特征见表1。
表 1 乳腺癌与他汀类药物相关研究变量的GWAS数据特征Table 1 Characteristics of GWAS data on study variables related to breast cancer and statinsPhenotypes/Genes Year Population Gender Sample size Sample source Data source Reference (PMID) HMGCR 2021 European Both 31 684 Whole blood eQTLGen 34475573 LDLR 2021 European Both 31 684 Whole blood eQTLGen 34475573 PCSK9 2020 European Both 73-670 Whole blood GTEx 32913098 ABCG8 2020 European Both 73-670 Cholesterol GTEx 32913098 APOB 2020 European Both 73-670 Cholesterol GTEx 32913098 NPC1L1 2020 European Both 73-670 Cholesterol GTEx 32913098 TC 2021 European Both 1 320 016 Whole blood GLGC 34887591 LDL-C 2021 European Both 1 320 016 Whole blood GLGC 34887591 Non-HDL-C 2021 European Both 1 320 016 Whole blood GLGC 34887591 All breast cancers 2017 European Both 228 951 - BCAC 29059683 ER+ breast cancer 2017 European Both 175 475 - BCAC 29059683 ER− breast cancer 2017 European Both 127 442 - BCAC 29059683 Notes: TC: total cholesterol; LDL-C: low-density lipoprotein cholesterol; non-HDL-C: non-high-density lipoprotein cholesterol; GLGC: Global Lipid Genetics Consortium; BCAC: Breast Cancer Association Consortium; ER: estrogen receptor; -: none. 1.2 胆固醇调控基因对乳腺癌发病风险影响的孟德尔随机化分析
由于研究暴露于六种胆固醇代谢调控基因的影响,因此预测暴露的工具变量为eQTL。从上述汇总数据中提取符合要求的eQTL,提取标准为:(1)工具变量必须相对常见,即最小等位基因频率大于1%;(2)工具变量与暴露的全基因组相关性P值小于5.0×10−8;(3)评估弱工具变量的F统计量大于10;(4)eQTL必须位于染色体上相应调控基因两侧100 kb的范围内。
采用基于汇总数据的孟德尔随机化(SMR)方法,评估与调控基因最显著相关的eQTL对全部乳腺癌、ER+乳腺癌及ER−乳腺癌发病风险的遗传影响。P<0.05提示遗传影响具有统计学意义[16]。通过工具变量异质性检验(Heterogeneity in dependent instrument, HEIDI )方法评估水平多效性,P>0.05提示相应分析不受水平多效性的影响。
上述分析均采用SMR软件(version 1.3.1)实施。六种胆固醇调控基因的名称、Ensembl编码、基因组序列、染色体(Chromosome, Chr)、基因定位(Location)及效应物见表2。
表 2 胆固醇调控基因的特征Table 2 Characterization of cholesterol-regulated genesGenes Ensembl code Sequence Chromosome Location Effectors HMGCR ENSG00000113161 GRCh37 05 74632993 –74657941 Cholesterol LDLR ENSG00000130164 GRCh37 19 11200139 –11244496 Cholesterol PCSK9 ENSG00000169174 GRCh37 01 55505221 –55530525 Cholesterol ABCG8 ENSG00000143921 GRCh37 02 44066110 –44110127 Cholesterol APOB ENSG00000084674 GRCh37 02 21224301 –21266945 Cholesterol NPC1L1 ENSG00000015520 GRCh37 07 44552134 –44580929 Cholesterol 1.3 相关基因调控下的胆固醇表型对乳腺癌发病风险影响的孟德尔随机化分析
因为暴露为相关基因调控下的胆固醇表型(TC、LDL-C及nonHDL-C),所以预测暴露的工具变量为SNP。符合要求的SNP亦从上述汇总数据中提取。提取标准为:(1)最小等位基因频率大于1%;(2)全基因组相关性P值小于5.0×10−8;(3)F统计量大于10;(4)以kb=100和R2=0.30为参数去除连锁不平衡;(5)工具变量必须在染色体上相应调控基因两侧100 kb以内。
采用传统的两样本孟德尔随机化法评估相关基因调控下的胆固醇表型对全部乳腺癌、ER+乳腺癌及ER−乳腺癌发病风险的遗传影响,这些方法包括随机效应模型的逆方差加权法(IVW)、MR-PRESSO法以及MR-Egger法。IVW是主要的两样本孟德尔随机化方法,具备最强的因果推断能力;MR-PRESSO可剔除工具变量中的潜在离群值,提供校正结果;MR-Egger方法最为保守,适用于工具变量质量较差的情况,能得到相对精确的结果。三种方法均报告比值比(OR)、95%CI和P值。P<0.05表示遗传影响具有统计学意义。如果三种方法结果不一致,要认定遗传影响是否具有统计学意义需同时满足以下两个条件:(1)IVW的P<0.05;(2)三种方法的OR值方向一致,即均大于1或均小于1。
采用Cochran’s Q检验和MR-Egger截距检验进行敏感性分析。其中,Cochran’s Q检验用于评估工具变量的异质性,而MR-Egger截距检验用于评估工具变量的水平多效性。所报告的P>0.05提示工具变量不受异质性或水平多效性的影响。
1.4 统计学方法
以上分析均采用装有Two-Sample-MR包的R软件(version 4.3.0)实施,检验水准为P<0.05。
2 结果
2.1 六种胆固醇代谢调控基因对乳腺癌发病风险的影响
调控基因HMGCR表达升高与全部乳腺癌及ER+乳腺癌的发病风险增加显著相关(全部乳腺癌:OR=1.112,95%CI: 1.003~1.234,P=0.044;ER+乳腺癌:OR=1.130,95%CI: 1.002~1.279,P=0.039)。而HMGCR表达与ER−乳腺癌的发病风险变化无显著相关性(P>0.05)。此外,其他五种调控基因的表达与全部乳腺癌、ER+乳腺癌及ER−乳腺癌的发病风险变化均无显著相关性(均P>0.05)。HEIDI检验结果显示,以上分析中未发现显著的水平多效性(均P>0.05),见表3。
表 3 六种胆固醇代谢调控基因对乳腺癌发病风险的影响Table 3 Effect of six cholesterol metabolism regulatory genes on breast cancer incidence riskPhenotypes/Genes Significant eQTL OR (95%CI) P (SMR) P (HEIDI) HMGCR All breast cancers rs6453133 1.112 (1.003−1.234) 0.044 0.850 ER+ breast cancer rs6453133 1.130 (1.002−1.279) 0.039 0.424 ER− breast cancer rs6453133 1.135 (0.939−1.372) 0.190 0.574 LDLR All breast cancers rs8110515 1.027 (0.845−1.247) 0.792 0.194 ER+ breast cancer rs8110515 1.019 (0.807−1.286) 0.876 0.514 ER− breast cancer rs8110515 0.908 (0.636−1.297) 0.597 0.581 PCSK9 All breast cancers rs472495 1.004 (0.932−1.081) 0.921 0.805 ER+ breast cancer rs472495 1.035 (0.946−1.131) 0.454 0.912 ER− breast cancer rs472495 0.931 (0.813−1.067) 0.307 0.862 ABCG8 All breast cancers rs78451356 0.986 (0.948−1.025) 0.470 - ER+ breast cancer rs78451356 0.987 (0.942−1.035) 0.594 - ER− breast cancer rs78451356 0.955 (0.889−1.027) 0.213 - APOB All breast cancers rs4665179 1.016 (0.984−1.050) 0.326 0.895 ER+ breast cancer rs4665179 0.996 (0.958−1.035) 0.843 0.802 ER− breast cancer rs4665179 1.043 (0.983−1.106) 0.168 0.155 NPC1L1 All breast cancers rs41279633 1.038 (0.998−1.079) 0.066 0.973 ER+ breast cancer rs41279633 1.044 (0.996−1.094) 0.073 0.727 ER− breast cancer rs41279633 0.999 (0.932−1.072) 0.981 0.759 Notes: eQTL: expression quantitative trait loci; SMR: Mendelian randomization based on summary data; HEIDI: heterogeneity in dependent instruments; −: none. 2.2 基因HMGCR调控下的外周胆固醇表型对乳腺癌发病风险的影响
在上述分析中,只有HMGCR基因显著影响乳腺癌的发病风险。因此,该调控基因被纳入后续分析,以确定HMGCR是否通过胆固醇途径影响乳腺癌发病风险。
IVW分析结果显示,在HMGCR调控下,外周血TC水平升高与全部乳腺癌及ER+乳腺癌的发病风险升高显著相关(全部乳腺癌:OR=1.214,95%CI: 1.113~1.324,P=1.160e-05;ER+乳腺癌:OR=1.209,95%CI: 1.090~1.340,P=3.181e-04)。MR-PRESSO的结果与IVW一致(P=6.235e-07,P=8.658e-05),而MR-Egger的结果方向与IVW相同。此外,IVW分析显示,在HMGCR的调控下,外周TC水平与ER−乳腺癌的发病风险无显著相关性(P=0.062)。在敏感性分析中,Cochran’s Q检验未发现显著的异质性(P>0.05),而MR-Egger截距检验未发现显著的水平多效性(P>0.05),见表4。
表 4 HMGCR调控下的外周血总胆固醇(TC)对乳腺癌发病风险的影响Table 4 Effect of HMGCR-regulated peripheral total cholesterol (TC) on breast cancer incidence riskBreast cancer types MR methods Nsnp OR (95% CI) P (MR) P (CO) P (MI) All breast cancers IVW 31 1.214 (1.113 – 1.324) 1.160e-05 0.992 0.847 MR-PRESSO 31 1.214 (1.143 – 1.290) 6.235e-07 - - MR-Egger 31 1.256 (0.886 – 1.779) 0.211 - - ER+ breast cancer IVW 31 1.209 (1.090 – 1.340) 3.181e-04 0.942 0.511 MR-PRESSO 31 1.209 (1.114 – 1.312) 8.658e-05 - - MR-Egger 31 1.385 (0.915 – 2.096) 0.134 - - ER− breast cancer IVW 31 1.163 (0.993 – 1.364) 0.062 0.996 0.653 MR-PRESSO 31 1.163 (1.046 – 1.293) 0.009 - - MR-Egger 31 1.008 (0.531 – 1.911) 0.981 - - Notes: CO: Cochran’s Q test; MI: MR-Egger intercept test; Nsnp: Number of SNP; −: none. IVW分析结果显示,在HMGCR的调控下,外周血LDL-C水平升高与全部乳腺癌及ER+乳腺癌的发病风险升高显著相关(全部乳腺癌:OR=1.198,95%CI: 1.105~1.300,P=1.248e-05;ER+乳腺癌:OR=1.199,95%CI: 1.089~1.321,P=2.231e-04)。MR-PRESSO的结果与IVW一致(P=6.019e-07,P=3.282e-05),而MR-Egger的结果方向与IVW相同。此外,IVW分析显示,在HMGCR的调控下,外周LDL-C水平与ER−乳腺癌的发病风险无显著相关性(P=0.133)。 在敏感性分析中,Cochran’s Q检验未发现显著的异质性(P>0.05),而MR-Egger截距检验未发现水平多效性(P>0.05),见表5。
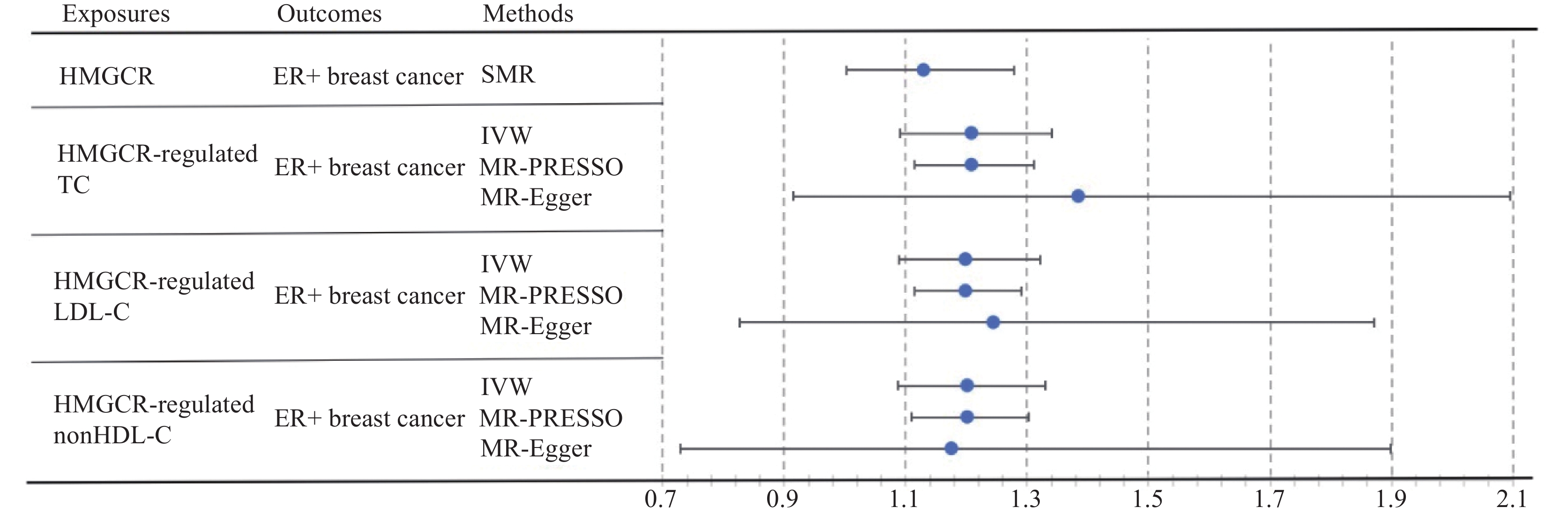
表 5 HMGCR调控下的外周血低密度脂蛋白胆固醇(LDL-C)对乳腺癌发病风险的影响Table 5 Effect of HMGCR-regulated peripheral low-density lipoprotein cholesterol (LDL-C) on breast cancer incidence riskBreast cancer types MR methods Nsnp OR (95% CI) P (MR) P (CO) P (MI) All breast cancers IVW 33 1.198 (1.105 – 1.300) 1.248e-05 0.992 0.905 MR-PRESSO 33 1.198 (1.132 – 1.269) 6.019e-07 - - MR-Egger 33 1.174 (0.833 – 1.655) 0.366 - - ER+ breast cancer IVW 33 1.199 (1.089 – 1.321) 2.231e-04 0.970 0.856 MR-PRESSO 33 1.199 (1.114 – 1.291) 3.282e-05 - - MR-Egger 33 1.245 (0.827 – 1.872) 0.302 - - ER− breast cancer IVW 33 1.121 (0.966 – 1.300) 0.133 0.995 0.669 MR-PRESSO 33 1.121 (1.012 – 1.241) 0.037 - - MR-Egger 33 0.979 (0.521 – 1.840) 0.948 - - Note: −: none. IVW分析结果显示,在HMGCR的调控下,外周血nonHDL-C水平升高与全部乳腺癌及ER+乳腺癌的发病风险升高显著相关(全部乳腺癌:OR=1.203,95%CI: 1.106~1.310,P=1.869e-05;ER+乳腺癌:OR=1.202,95%CI: 1.087~1.330,P=3.520e-04)。MR-PRESSO的结果与IVW一致(P=2.146e-06,P=1.388e-04),而MR-Egger的结果方向与IVW相同。此外,IVW分析显示,在HMGCR的调控下,外周nonHDL-C水平与ER−乳腺癌的发病风险无显著相关性(P=0.055)。在敏感性分析中,Cochran’s Q检验未发现显著的异质性(P>0.05),而MR-Egger截距检验未发现显著的水平多效性(P>0.05),见表6。HMGCR以及其调控的胆固醇表型对ER+乳腺癌发病风险影响见图1。
表 6 HMGCR调控下的外周非高密度脂蛋白胆固醇(non-HDL-C)对乳腺癌发病风险的影响Table 6 Effect of HMGCR-regulated peripheral non-high-density lipoprotein cholesterol (non-HDL-C) on breast cancer incidence riskBreast cancer types MR Methods Nsnp OR (95%CI) P (MR) P (CO) P (MI) All breast cancers IVW 25 1.203 (1.106 – 1.310) 1.869e-05 0.985 0.921 MR-PRESSO 25 1.203 (1.135 – 1.276) 2.146e-06 - - MR-Egger 25 1.179 (0.788 – 1.765) 0.431 - - ER+breast cancer IVW 25 1.202 (1.087 – 1.330) 3.520e-04 0.922 0.927 MR-PRESSO 25 1.202 (1.110 – 1.302) 1.388e-04 - - MR-Egger 25 1.176 (0.728 – 1.899) 0.515 - - ER− breast cancer IVW 25 1.164 (0.997 – 1.360) 0.055 0.990 0.959 MR-PRESSO 25 1.164 (1.049 – 1.293) 0.009 - - MR-Egger 25 1.187 (0.565 – 2.492) 0.655 - - Note: −: none. 3 讨论
近年来,医学界在乳腺癌致病因素和发病机制的研究取得显著进展,各种新型治疗手段不断涌现,显著降低了其死亡率,改善了患者预后[17-18]。然而,乳腺癌的致病因素和发病机制仍有许多未知领域亟待探索。在此背景下,本研究通过药靶孟德尔随机化方法,在基因水平上探讨他汀类药物对乳腺癌发病风险的影响,旨在填补乳腺癌发病机制中胆固醇代谢相关的科研空白,深化对乳腺癌发生和发展的认识,助力乳腺癌的预防与控制。
本研究结果发现,HMGCR基因表达水平每升高一个标准差,全部乳腺癌和ER+乳腺癌的发病风险增加约10%。作为HMGCR抑制剂,他汀类药物在基因水平上对全部乳腺癌和ER+乳腺癌具有保护作用。鉴于他汀类药物具有降脂、抗炎和抗氧化应激等多种生物学作用,进一步探讨HMGCR调控下胆固醇表型对乳腺癌发病风险的影响。结果显示,HMGCR调控下的外周血TG、LDL-C和nonHDL-C水平每增加一个标准差,全部乳腺癌和ER+乳腺癌的发病风险增加约20%。这初步表明,他汀类药物对乳腺癌的保护作用部分源于其降低胆固醇水平的生物学效应。
Murto等的一项大型队列研究进一步支持了他汀类药物在13 378名乳腺癌患者中的潜在益处,特别是在降低乳腺癌特异性死亡率方面。该研究发现,使用他汀类药物与降低乳腺癌死亡风险相关,尤其是对于雌激素受体阳性(ER+)患者[19],这与本研究结果相符。近期,Watson等汇总2007年至2022年间发表的26项关于他汀类药物治疗乳腺癌的体内实验,显示乳腺癌小鼠模型中,单独使用他汀类药物或与抗癌疗法联合使用均显示出抗肿瘤作用,其效果与药物的使用时间、剂量和乳腺癌亚型等因素相关[20]。O'Grady等的体外实验则证实,他汀类药物与多柔比星/多西紫杉醇联合使用,可有效抑制乳腺癌细胞的增殖并诱导其凋亡,p53突变状态可作为他汀类药物反应的潜在预测生物标志物[21]。以上基础研究结果均支持本研究发现。
本研究采用两种药靶孟德尔随机化方法。第一种是SMR方法,用于评估特定基因表达与表型间的因果关系。第二种方法基于传统的两样本孟德尔随机化方法,选择特定基因的效应物作为暴露因素,工具变量位于染色体上该基因的100 kb范围内,从而暴露因素可理解为特定基因调控下的效应物水平。通过逆向思维,该方法也可用于证实特定基因通过何种效应物调控结局因素,实现初步的机制分析。
另外,本研究未能证实他汀类药物使用与ER−乳腺癌发病风险变化间存在显著相关性。他汀类药物对ER+乳腺癌有保护作用而对ER−患者无显著影响,其可能机制有如下几方面:(1)抑制胆固醇合成途径:他汀类药物通过抑制HMG-CoA还原酶,阻断甲羟戊酸途径,降低胆固醇及异戊二烯类产物的合成,这些产物对于细胞膜的完整性、信号转导和蛋白质修饰至关重要,其减少可能影响肿瘤细胞的增殖和存活[22]。(2)干扰雌激素受体信号转导:他汀类药物可能直接影响ERα的表达和活性。Goda等发现,瑞舒伐他汀与尼洛替尼联用可下调ERα蛋白表达,抑制ERα下游目标基因C3和pS2的活性,从而抑制ER+乳腺癌细胞的增殖[23]。(3)诱导细胞凋亡和阻滞细胞周期:围手术期应用他汀类药物增加凋亡相关蛋白(如caspase-3)的表达,促进肿瘤细胞凋亡;影响细胞周期调控蛋白(如Cyclin D1、p27),阻滞细胞周期进程[24]。基于上述机制分析,他汀类药物在乳腺癌预防和治疗中具有潜在的应用价值,特别是对于ER+乳腺癌患者。建议在临床实践中,应考虑定期监测乳腺癌患者的胆固醇水平,并评估他汀类药物的应用。
此外,本研究未发现其他五种胆固醇调控基因(LDLR、PCSK9、ABCG8、APOB及NPC1L1)对乳腺癌发病风险有显著影响。然而,这些基因在此前研究中被证实具有明确的胆固醇调控作用[10]。这一结果从侧面提示,HMGCR对乳腺癌的保护机制可能不仅依赖于其降胆固醇作用。这一推测与前述他汀类药物的抗乳腺癌机制可能涉及雌激素的假设相一致。值得注意的是,胆固醇本身参与雌激素的代谢,胆固醇机制和雌激素机制可能存在交互作用。
本研究存在如下不足:首先,仅探讨了六种胆固醇调控基因,未纳入更多已知或未知的相关基因,以及主要调控外周甘油三酯的基因,导致对胆固醇或血脂代谢对乳腺癌发病风险影响的探讨不够全面;其次,对于HMGCR降低乳腺癌发病风险的潜在机制,仅进行了初步分析和推测,尚未直接证实其病理生理过程;第三,研究对象仅限于欧洲人群,未包含亚洲或非洲人群的数据。这些不足主要源于数据库中相关汇总数据的缺乏,无法支撑更全面的研究。因此,待未来相关数据库充实和完善后,将对其进行深入探讨。同时应开展大样本随机对照临床试验,以验证他汀类药物在乳腺癌预防和治疗中的有效性和安全性。这将有助于制定具体的临床指导和干预策略,为ER+乳腺癌患者提供新的治疗选择。
综上,本研究证实他汀类药物在基因水平上可降低ER+乳腺癌的发病风险,而对ER−乳腺癌则无类似保护作用。推测他汀类药物对乳腺癌的保护机制可能涉及胆固醇代谢、雌激素代谢等多种病理生理过程。这些结果有助于深刻理解胆固醇代谢在乳腺癌发病机制中作用,并为开发新的抗乳腺癌治疗策略提供初步理论依据。
Competing interests: The authors declare that they have no competing interests.利益冲突声明:所有作者均声明不存在利益冲突。作者贡献:胡 荻:思路设计、数据分析、制图、论文撰写水一方:结果验证、论文审阅修改苗轲轲:数据获取、材料收集李孟圈:资金支持、课题指导、论文审阅修改 -
表 1 乳腺癌与他汀类药物相关研究变量的GWAS数据特征
Table 1 Characteristics of GWAS data on study variables related to breast cancer and statins
Phenotypes/Genes Year Population Gender Sample size Sample source Data source Reference (PMID) HMGCR 2021 European Both 31 684 Whole blood eQTLGen 34475573 LDLR 2021 European Both 31 684 Whole blood eQTLGen 34475573 PCSK9 2020 European Both 73-670 Whole blood GTEx 32913098 ABCG8 2020 European Both 73-670 Cholesterol GTEx 32913098 APOB 2020 European Both 73-670 Cholesterol GTEx 32913098 NPC1L1 2020 European Both 73-670 Cholesterol GTEx 32913098 TC 2021 European Both 1 320 016 Whole blood GLGC 34887591 LDL-C 2021 European Both 1 320 016 Whole blood GLGC 34887591 Non-HDL-C 2021 European Both 1 320 016 Whole blood GLGC 34887591 All breast cancers 2017 European Both 228 951 - BCAC 29059683 ER+ breast cancer 2017 European Both 175 475 - BCAC 29059683 ER− breast cancer 2017 European Both 127 442 - BCAC 29059683 Notes: TC: total cholesterol; LDL-C: low-density lipoprotein cholesterol; non-HDL-C: non-high-density lipoprotein cholesterol; GLGC: Global Lipid Genetics Consortium; BCAC: Breast Cancer Association Consortium; ER: estrogen receptor; -: none. 表 2 胆固醇调控基因的特征
Table 2 Characterization of cholesterol-regulated genes
Genes Ensembl code Sequence Chromosome Location Effectors HMGCR ENSG00000113161 GRCh37 05 74632993 –74657941 Cholesterol LDLR ENSG00000130164 GRCh37 19 11200139 –11244496 Cholesterol PCSK9 ENSG00000169174 GRCh37 01 55505221 –55530525 Cholesterol ABCG8 ENSG00000143921 GRCh37 02 44066110 –44110127 Cholesterol APOB ENSG00000084674 GRCh37 02 21224301 –21266945 Cholesterol NPC1L1 ENSG00000015520 GRCh37 07 44552134 –44580929 Cholesterol 表 3 六种胆固醇代谢调控基因对乳腺癌发病风险的影响
Table 3 Effect of six cholesterol metabolism regulatory genes on breast cancer incidence risk
Phenotypes/Genes Significant eQTL OR (95%CI) P (SMR) P (HEIDI) HMGCR All breast cancers rs6453133 1.112 (1.003−1.234) 0.044 0.850 ER+ breast cancer rs6453133 1.130 (1.002−1.279) 0.039 0.424 ER− breast cancer rs6453133 1.135 (0.939−1.372) 0.190 0.574 LDLR All breast cancers rs8110515 1.027 (0.845−1.247) 0.792 0.194 ER+ breast cancer rs8110515 1.019 (0.807−1.286) 0.876 0.514 ER− breast cancer rs8110515 0.908 (0.636−1.297) 0.597 0.581 PCSK9 All breast cancers rs472495 1.004 (0.932−1.081) 0.921 0.805 ER+ breast cancer rs472495 1.035 (0.946−1.131) 0.454 0.912 ER− breast cancer rs472495 0.931 (0.813−1.067) 0.307 0.862 ABCG8 All breast cancers rs78451356 0.986 (0.948−1.025) 0.470 - ER+ breast cancer rs78451356 0.987 (0.942−1.035) 0.594 - ER− breast cancer rs78451356 0.955 (0.889−1.027) 0.213 - APOB All breast cancers rs4665179 1.016 (0.984−1.050) 0.326 0.895 ER+ breast cancer rs4665179 0.996 (0.958−1.035) 0.843 0.802 ER− breast cancer rs4665179 1.043 (0.983−1.106) 0.168 0.155 NPC1L1 All breast cancers rs41279633 1.038 (0.998−1.079) 0.066 0.973 ER+ breast cancer rs41279633 1.044 (0.996−1.094) 0.073 0.727 ER− breast cancer rs41279633 0.999 (0.932−1.072) 0.981 0.759 Notes: eQTL: expression quantitative trait loci; SMR: Mendelian randomization based on summary data; HEIDI: heterogeneity in dependent instruments; −: none. 表 4 HMGCR调控下的外周血总胆固醇(TC)对乳腺癌发病风险的影响
Table 4 Effect of HMGCR-regulated peripheral total cholesterol (TC) on breast cancer incidence risk
Breast cancer types MR methods Nsnp OR (95% CI) P (MR) P (CO) P (MI) All breast cancers IVW 31 1.214 (1.113 – 1.324) 1.160e-05 0.992 0.847 MR-PRESSO 31 1.214 (1.143 – 1.290) 6.235e-07 - - MR-Egger 31 1.256 (0.886 – 1.779) 0.211 - - ER+ breast cancer IVW 31 1.209 (1.090 – 1.340) 3.181e-04 0.942 0.511 MR-PRESSO 31 1.209 (1.114 – 1.312) 8.658e-05 - - MR-Egger 31 1.385 (0.915 – 2.096) 0.134 - - ER− breast cancer IVW 31 1.163 (0.993 – 1.364) 0.062 0.996 0.653 MR-PRESSO 31 1.163 (1.046 – 1.293) 0.009 - - MR-Egger 31 1.008 (0.531 – 1.911) 0.981 - - Notes: CO: Cochran’s Q test; MI: MR-Egger intercept test; Nsnp: Number of SNP; −: none. 表 5 HMGCR调控下的外周血低密度脂蛋白胆固醇(LDL-C)对乳腺癌发病风险的影响
Table 5 Effect of HMGCR-regulated peripheral low-density lipoprotein cholesterol (LDL-C) on breast cancer incidence risk
Breast cancer types MR methods Nsnp OR (95% CI) P (MR) P (CO) P (MI) All breast cancers IVW 33 1.198 (1.105 – 1.300) 1.248e-05 0.992 0.905 MR-PRESSO 33 1.198 (1.132 – 1.269) 6.019e-07 - - MR-Egger 33 1.174 (0.833 – 1.655) 0.366 - - ER+ breast cancer IVW 33 1.199 (1.089 – 1.321) 2.231e-04 0.970 0.856 MR-PRESSO 33 1.199 (1.114 – 1.291) 3.282e-05 - - MR-Egger 33 1.245 (0.827 – 1.872) 0.302 - - ER− breast cancer IVW 33 1.121 (0.966 – 1.300) 0.133 0.995 0.669 MR-PRESSO 33 1.121 (1.012 – 1.241) 0.037 - - MR-Egger 33 0.979 (0.521 – 1.840) 0.948 - - Note: −: none. 表 6 HMGCR调控下的外周非高密度脂蛋白胆固醇(non-HDL-C)对乳腺癌发病风险的影响
Table 6 Effect of HMGCR-regulated peripheral non-high-density lipoprotein cholesterol (non-HDL-C) on breast cancer incidence risk
Breast cancer types MR Methods Nsnp OR (95%CI) P (MR) P (CO) P (MI) All breast cancers IVW 25 1.203 (1.106 – 1.310) 1.869e-05 0.985 0.921 MR-PRESSO 25 1.203 (1.135 – 1.276) 2.146e-06 - - MR-Egger 25 1.179 (0.788 – 1.765) 0.431 - - ER+breast cancer IVW 25 1.202 (1.087 – 1.330) 3.520e-04 0.922 0.927 MR-PRESSO 25 1.202 (1.110 – 1.302) 1.388e-04 - - MR-Egger 25 1.176 (0.728 – 1.899) 0.515 - - ER− breast cancer IVW 25 1.164 (0.997 – 1.360) 0.055 0.990 0.959 MR-PRESSO 25 1.164 (1.049 – 1.293) 0.009 - - MR-Egger 25 1.187 (0.565 – 2.492) 0.655 - - Note: −: none. -
[1] Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2024, 74(3): 229-263. doi: 10.3322/caac.21834
[2] 胡志强, 游伟程, 潘凯枫, 等. 中、美两国癌症流行特征分析——《2023美国癌症统计报告》解读[J]. 科技导报, 2023, 41(18): 18-28. [Hu ZQ, You WC, Pan KF, et al. Epidemiological characteristics of cancers in China and America: Interpretation of the report of American cancer statistics, 2023[J]. Ke Ji Dao Bao, 2023, 41(18): 18-28.] Hu ZQ, You WC, Pan KF, et al. Epidemiological characteristics of cancers in China and America: Interpretation of the report of American cancer statistics, 2023[J]. Ke Ji Dao Bao, 2023, 41(18): 18-28.
[3] Lei S, Zheng R, Zhang S, et al. Breast cancer incidence and mortality in women in China: temporal trends and projections to 2030[J]. Cancer Biol Med, 2021, 18(3): 900-909. doi: 10.20892/j.issn.2095-3941.2020.0523
[4] Waks AG, Winer EP. Breast Cancer Treatment: A Review[J]. JAMA, 2019, 321(3): 288-300. doi: 10.1001/jama.2018.19323
[5] Narii N, Zha L, Komatsu M, et al. Cholesterol and breast cancer risk: a cohort study using health insurance claims and health checkup databases[J]. Breast Cancer Res Treat, 2023, 199(2): 315-322. doi: 10.1007/s10549-023-06917-z
[6] Wang Y, Liu F, Sun L, et al. Association between human blood metabolome and the risk of breast cancer[J]. Breast Cancer Res, 2023, 25(1): 9. doi: 10.1186/s13058-023-01609-4
[7] Ben Hassen C, Goupille C, Vigor C, et al. Is cholesterol a risk factor for breast cancer incidence and outcome?[J]. J Steroid Biochem Mol Biol, 2023, 232: 106346. doi: 10.1016/j.jsbmb.2023.106346
[8] Ricco N, Kron SJ. Statins in Cancer Prevention and Therapy[J]. Cancers (Basel), 2023, 15(15): 3948. doi: 10.3390/cancers15153948
[9] Zhao G, Ji Y, Ye Q, et al. Effect of statins use on risk and prognosis of breast cancer: a meta-analysis[J]. Anticancer Drugs, 2022, 33(1): e507-e518. doi: 10.1097/CAD.0000000000001151
[10] Abdul-Rahman T, Bukhari SMA, Herrera EC, et al. Lipid Lowering Therapy: An Era Beyond Statins[J]. Curr Probl Cardiol, 2022, 47(12): 101342. doi: 10.1016/j.cpcardiol.2022.101342
[11] Miziak P, Baran M, Błaszczak E, et al. Estrogen Receptor Signaling in Breast Cancer[J]. Cancers (Basel), 2023, 15(19): 4689. doi: 10.3390/cancers15194689
[12] Võsa U, Claringbould A, Westra HJ, et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression[J]. Nat Genet, 2021, 53(9): 1300-1310. doi: 10.1038/s41588-021-00913-z
[13] GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues[J]. Science, 2020, 369(6509): 1318-1330.
[14] Graham SE, Clarke SL, Wu KH, et al. The power of genetic diversity in genome-wide association studies of lipids[J]. Nature, 2021, 600(7890): 675-679. doi: 10.1038/s41586-021-04064-3
[15] Michailidou K, Lindström S, Dennis J, et al. Association analysis identifies 65 new breast cancer risk loci[J]. Nature, 2017, 551(7678): 92-94. doi: 10.1038/nature24284
[16] Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets[J]. Nat Genet, 2016, 48(5): 481-487. doi: 10.1038/ng.3538
[17] 刘佳, 周星彤, 孙强. 多灶性/多中心性乳腺癌研究进展[J]. 协和医学杂志, 2024, 15(3): 632-641. [Liu J, Zhou XT, Sun Q. Research Progress of Multifocal/Multicentric Breast Cancer[J]. Xie He Yi Xue Za Zhi, 2024, 15(3): 632-641.] doi: 10.12290/xhyxzz.2023-0562 Liu J, Zhou XT, Sun Q. Research Progress of Multifocal/Multicentric Breast Cancer[J]. Xie He Yi Xue Za Zhi, 2024, 15(3): 632-641. doi: 10.12290/xhyxzz.2023-0562
[18] 何国珍, 刘晓, 员晓云, 等. 中医药防治乳腺癌相关信号通路的研究进展[J/OL]. 中华中医药学刊: 1-19 [2024-05-06]. http://kns.cnki.net/kcms/detail/21.1546.R.20240506.1130.002.html. [He GZ, Liu X, Yun XY, et al. Research progress of signaling pathways related to the prevention and treatment of breast cancer by traditional Chinese medicine[J/OL]. Zhonghua Zhong Yi Yao Xue Kan: 1-19 [2024-05-06]. http://kns.cnki.net/kcms/detail/21.1546.R.20240506.1130.002.html.] He GZ, Liu X, Yun XY, et al. Research progress of signaling pathways related to the prevention and treatment of breast cancer by traditional Chinese medicine[J/OL]. Zhonghua Zhong Yi Yao Xue Kan: 1-19 [2024-05-06]. http://kns.cnki.net/kcms/detail/21.1546.R.20240506.1130.002.html.
[19] Murto MO, Simolin N, Arponen O, et al. Statin Use, Cholesterol Level, and Mortality Among Females With Breast Cancer[J]. JAMA Netw Open, 2023, 6(11): e2343861. doi: 10.1001/jamanetworkopen.2023.43861
[20] Watson R, Tulk A, Erdrich J. The Link Between Statins and Breast Cancer in Mouse Models: A Systematic Review[J]. Cureus, 2022, 14(11): e31893.
[21] O'Grady S, Crown J, Duffy MJ. Statins inhibit proliferation and induce apoptosis in triple-negative breast cancer cells[J]. Med Oncol, 2022, 39(10): 142. doi: 10.1007/s12032-022-01733-9
[22] Bjarnadottir O, Romero Q, Bendahl PO, et al. Targeting HMG-CoA reductase with statins in a window-of-opportunity breast cancer trial[J]. Breast Cancer Res Treat, 2013, 138(2): 499-508. doi: 10.1007/s10549-013-2473-6
[23] Goda AE, Elsisi AE, Sokkar SS, et al. Enhanced in vivo targeting of estrogen receptor alpha signaling in murine mammary adenocarcinoma by nilotinib/rosuvastatin novel combination[J] Toxicol Appl Pharmacol, 2020, 404: 115185.
[24] Kamal A, Boerner J, Assad H, et al. The Effect of Statins on Markers of Breast Cancer Proliferation and Apoptosis in Women with In Situ or Early-Stage Invasive Breast Cancer[J]. Int J Mol Sci, 2024, 25(17): 9587. doi: 10.3390/ijms25179587



 下载:
下载: