-
摘要:目的
分析11株来自中国尚未明确分型的HAdV病毒株完整基因组的基因特征并分型。
方法从GenBank数据库中下载到67条已明确基因型的HAdV基因组序列和7条HAdV参照基因组序列,将这74条参照序列与11株未明确分型的HAdV病毒株基因组序列进行基因结构、基因同源性以及系统进化等分析,从而确定其分型。
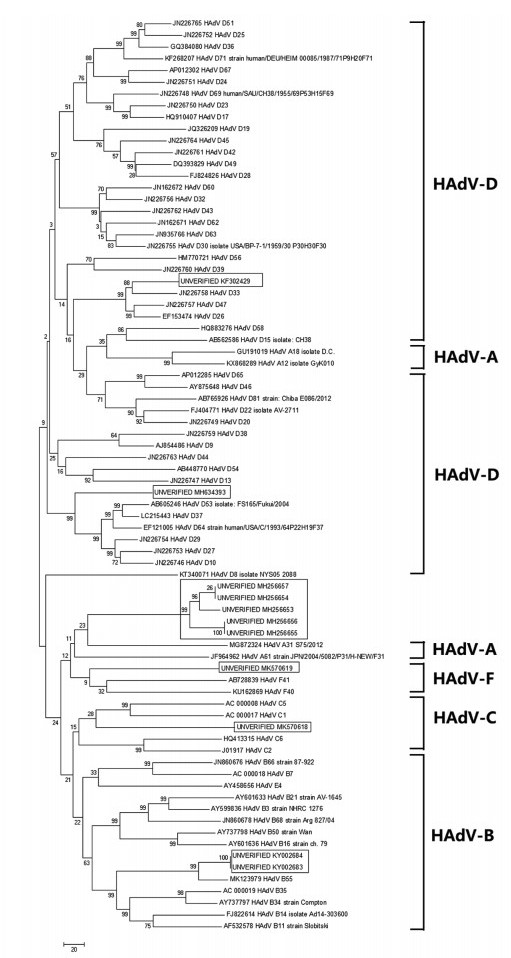
结果从系统进化分析结果预测出6株HAdV可能的分型,再由多序列比对分析进行验证。其中两株HAdV通过多序列比对结果发现与HAdV-B参照基因组序列之间的同源性为97.8%,属于B型HAdV;两株HAdV与HAdV-D参照基因组序列之间的同源性为92.8%,属于D型HAdV;还有两株HAdV的多序列比对结果与系统进化分析预测结果不一致,无法推断其分型。剩余5株未能预测的HAdV通过与7条HAdV参照序列进行多序列比对分析,发现与B型HAdV基因组序列的同源性达到了97.9%,推测这5株HAdV基因组可能属于B型HAdV。
结论从基因角度来对11株未知的HAdV基因组进行分型,预测有7株可能属于B型HAdV,有两株可能属于D型HAdV,还有两株无法判断其分型。其中7株B型HAdV可能适合作为溶瘤腺病毒进行骨肉瘤的治疗。
Abstract:ObjectiveTo analyze the type and gene features of 11 unverified HAdVs in China.
MethodsWe downloaded HAdV genome sequences of 67 strains with clear genotypes and the HAdV reference genome sequences of 7 strains from GenBank database, analyzed them together with eleven unverified HAdV gene sequences and classified the species of each sequence based on the genetic structure, sequence similarity and phylogenetic analysis.
ResultsPhylogenetic analysis results predicted the classification of six sequences: two of them belonged to HAdV-B and two belonged to HAdV-D (the gene homology were 97.8% and 92.8% in comparison with reference gene respectively), the rest of two sequences were not conclusive. Multiple sequence alignment was used to predict the species of five HAdVs. All of them belonged to species HAdV-B, the sequence similarity compared with reference gene was 97.9%.
ConclusionGenotyping of the unverified HAdV genome from a genetic point of view predicts that seven strains might belong to species HAdV-B, two strains might belong to species HAdV-D and two strains could not be classified. The seven HAdV-B strains might be suitable for the treatment of osteosarcoma as oncolytic adenovirus.
-
Key words:
- HAdV /
- Phylogenetic analysis /
- Oncolytic adenovirus /
- Classification /
- Cancer
-
0 引言
乳腺癌(breast cancer, BC)是全世界女性最常见的癌症之一,其发病率处于持续增长状态。自20世纪90年代以来,中国乳腺癌发病率的增长速度是全球的两倍多,同时也是中国女性第六大癌症死因[1]。此外,世界大多数地区甲状腺癌(thyroid cancer, TC)的发病率也呈现出上升趋势,且已成为女性第八位最常见的恶性肿瘤,而我国甲状腺癌新发病例数占全球新发病例数的比例高达15.6%[2]。这两种疾病发病率的增加使我们越来越多的关注到两者并发的存在。
在过去的十几年中,已有多项研究提供了可靠的证据,表明乳腺癌与甲状腺癌之间存在潜在的、双向的致病关系[3]。Van等[4]对美国癌症协会的数据进行了评估,发现女性甲状腺癌患者随后发生乳腺癌的风险增加了0.67倍,女性乳腺癌患者的甲状腺癌发病率则增加了两倍,而男性患有甲状腺癌后续发生乳腺癌的风险增加了29倍,男性乳腺癌患者其后甲状腺癌的发病率则增加了19倍。Huang等[5]在调查中也证实并量化了两种癌症并发增加的存在,且表示仅监测偏倚不能完全解释甲状腺癌与乳腺癌之间的关系,两者之间可能存在共同的病理生理因素。部分研究显示BC合并TC有其独特的临床病理特征,且在BC发生后的3年内TC的患病风险很高[6]。因此,了解甲状腺癌和乳腺癌之间的并发关系对于两种疾病的临床监测和管理都具有重要意义。本文就原发性乳腺癌合并甲状腺癌的流行病学证据和可能的机制作一综述。
1 遗传因素
在癌症的发展中,家族联系是很重要的因素。一项针对瑞典患者的研究发现,患有乳腺癌的女性其一级亲属患甲状腺癌的风险增加[7],同时在美国人口中也观察到了BC与TC的高度家族关联[8]。遗传因素在乳腺肿瘤和甲状腺肿瘤的并发中成为了最有力的解释,因为驱动两种癌症的许多基因突变都相似。目前已知考登综合征(cowden syndrome, CS)是一种由于抑癌基因PTEN突变所导致的罕见常染色体显性遗传病,研究发现CS增加了同一个体罹患乳腺癌和甲状腺癌的风险,但这种突变在甲状腺癌与乳腺癌同时并发的非CS患者中很少见到。
1.1 PARP4突变
最近的一项研究确定了PARP4的种系突变是原发性甲状腺癌和乳腺癌的易感因素[3],参与调查的14例患者中有6例(43%)发现了PARP4的G496V和T1170I变体,而在对照组中它们的频率仅为0.5%。PARP4属于聚ADP-核糖聚合酶(poly ADP-ribose polymerase, PARP)家族成员[9],PARP家族共17个成员,是DNA修复、转录调控和RNA干扰的重要组成部分,由于PARP4含有BRCA1羧基末端(BRCT)域重复序列,BRCT可以与磷酸化的DNA损伤敏感蛋白结合,进而将PARPs募集到DNA损伤位点,因此我们猜测PARP4是DNA修复的关键组成部分。根据先前的报道[10],一些DNA修复基因在甲状腺乳头状癌、乳腺癌、卵巢癌以及很多同时性癌症中都发生了明显突变。
1.2 RET/PTC激酶
RET/PTC激酶最早作为甲状腺乳头状癌中最常见的致癌基因为人们所熟知,其突变和重排促进了遗传性甲状腺癌的发展[11]。最近它被发现是一种雌激素依赖性的酪氨酸激酶[12],在乳腺癌细胞中,雌激素与雌激素受体结合,随后结合体与雌激素受体叉头框蛋白A1被募集到雌激素反应元件和叉头反应元件位点,两个转录因子相互作用加强RET/PTC激酶基因的转录,使RET/PTC激酶表达上调,当胶质细胞源性神经营养因子与RET相互作用后,将引起RET自身的磷酸化,随后增强下游的细胞外调节蛋白激酶、蛋白激酶B和p38丝裂原活化蛋白激酶的磷酸化作用,促进乳腺癌细胞增殖,见图 1。因此,RET/PTC激酶的过表达与ER阳性乳腺癌的不良预后有关。
![]() 图 1 RET/PTC在乳腺癌中的作用FOXA1: ER-forkhead box protein A1; FHRE: forkhead responsive element; ERE: estrogen response element; GDNF: glial cell line-derived neurotrophic factor; ERK: extracellular regulated protein kinases; AKT: protein kinase B; P38: 38kD mitogen-activated protein kinase.Figure 1 Role of RET/PTC in breast cancer
图 1 RET/PTC在乳腺癌中的作用FOXA1: ER-forkhead box protein A1; FHRE: forkhead responsive element; ERE: estrogen response element; GDNF: glial cell line-derived neurotrophic factor; ERK: extracellular regulated protein kinases; AKT: protein kinase B; P38: 38kD mitogen-activated protein kinase.Figure 1 Role of RET/PTC in breast cancer2 钠碘同向转运体
2.1 钠碘同向转运体与乳腺癌
钠碘同向转运体(sodium iodide symporter, NIS)是一种介导碘摄取的跨膜糖蛋白,在甲状腺组织以及哺乳期乳腺组织中高表达[13]。早在1974年Eskin等[14]就证明了BC组织可以吸收放射性碘,而在正常的、非泌乳的乳腺组织中不会发生摄取。随后研究人员通过对患有浸润性导管癌的妇女进行研究,证实了NIS可以在转录和翻译水平上表达,且在癌组织中具有转运碘的能力[15]。与NIS mRNA表达相比,碘化物的摄取水平却相对较低。Wapnir等[16]针对这一问题展开了研究,并发现NIS蛋白位于泌乳乳腺的基底外侧膜上,而在BC中,细胞内和质膜均观察到了NIS的表达,但其功能活性和调节定位的分子机制仍不清楚,目前的研究中也尚未进行评估,一种可能性是NIS的翻译后修饰可调节NIS的细胞内分布和活性。因此,人们猜测NIS蛋白的错误定位可能导致了NIS表达水平与所观察到的碘摄取之间存在差异。同时,根据内分泌学的研究显示,无论是碘的缺乏还是摄入过量都会在BC中起不利作用[17-18]。
2.2 钠碘同向转运体与甲状腺癌
研究人员对于NIS在不同分化类型的甲状腺癌中的表达已进行了广泛的研究,据报道未分化型甲状腺癌的细胞膜上NIS不表达或低表达,但关于分化型的乳头状癌和滤泡状癌中NIS表达的意见尚未统一,部分结果显示,相比正常甲状腺组织,甲状腺乳头状癌(papillary thyroid carcinoma, PTC)中的NIS表达明显降低[19],从而导致聚碘能力下降。另有报道则表明NIS在分化型甲状腺癌(differentiated thyroid cancer, DTC)中表达增加,但其多数定位于细胞质中[16]。实际上在正常的甲状腺组织中,NIS蛋白主要在基底外侧膜上表达,因此癌组织仍然无法有效摄取碘。碘缺乏是目前公认的导致TC的危险因素之一,在正常甲状腺细胞中,缺碘会诱导血管内皮生长因子(vascular endothelial growth factor, VEGF)mRNA暂时性激活从而保护甲状腺细胞功能,但在TC细胞中,VEGF mRNA的表达持续时间显著延长。因此,碘的缺乏可能有助于血管生成转换,从而为TC的发生提供了血管微环境[20]。
综上所述,NIS在甲状腺癌和乳腺癌中主要为细胞质表达,NIS定位不当所介导的碘的摄取异常可能在BC和TC的致癌因素中起作用。所以,有关诱导NIS定位转化、调节功能性NIS在细胞内的表达,以增加放射性碘疗法的疗效在BC合并TC的患者中展现出广阔的前景。
3 雌激素与雌激素受体
雌激素致癌的重要机制之一就是刺激组织的生长和抑制凋亡,目前,已有广泛的研究报道雌激素在BC和前列腺癌中的作用[21]。无论是内源性雌激素水平的持续升高还是给予外源性雌激素治疗都会增加乳腺癌的发病率,主要致病机制如下:雌激素通过与雌激素受体(estrogen receptor, ER)结合使ER构象发生改变,形成的二聚体与雌激素反应元件(ERE)相互作用,调控下游基因的表达,并参与细胞的增殖和分化,促进乳腺癌的发生,增强了乳腺癌细胞的运动,同时诱导表达ER的乳腺癌细胞向远处器官或淋巴结侵袭和转移[22]。此外,雌激素的氧化代谢产物可以直接或间接促进乳腺癌的发生和发展,雌激素通过CYP1B1基因催化4-羟基化反应产生的4-羟基儿茶酚雌激素(4-OHE)具有致癌作用,随后进一步代谢,与DNA共价结合,形成不稳定加合物,引发脱嘌呤突变促进BC的发生与发展[21]。
女性甲状腺肿瘤的发病率是男性的3倍[23],而男性与女性之间最大的差异就是性激素的水平,雌激素对于女性的多种器官都具有广泛的影响,据报道,在绝经后需使用雌激素治疗妇科疾病但未进行低剂量激素替代治疗的妇女,其发生甲状腺肿瘤的风险也较高[22]。这些流行病学研究指出了雌激素在甲状腺肿瘤中的致病作用。研究人员在大鼠的甲状腺细胞中证实了17β-雌二醇具有直接的生长刺激作用。用17β-雌二醇刺激甲状腺良恶性细胞,可导致增殖速率提高和细胞周期蛋白D1表达增强,在恶性肿瘤细胞中,D1在3 h后即可发生最大程度的表达,而在良性细胞中,D1达到峰值的时间显著延迟[24]。D1在调控细胞周期G1/S转变中起关键作用,其过度表达将会导致细胞失控、癌变。此外,Vannucchi等[25]的研究所示,雌激素受体在正常和恶性甲状腺组织中均表达,其中,甲状腺癌组织的受体水平高于正常甲状腺组织,并且与更高的局部转移率相关。膜结合的ER与丝裂原活化蛋白激酶(mitogen-activated protein kinase, MAPK)和磷脂酰肌醇3激酶(phosphatidylinositol 3-kinase, PI3K)信号转导途径相连,这些途径在PTC中可以被酪氨酸激酶受体TRK和RET/PTC基因的重排或BRAF突变激活[26],雌激素通过膜结合的ER协同刺激MAPK和PI3K通路,见图 2。因此,女性中TC的高发生率可能归因于功能性ER的表达,该功能性ER有助于增强甲状腺细胞的有丝分裂,并参与迁移和侵袭能力的细胞过程。这些发现将促进针对肿瘤侵袭和迁移的抗雌激素疗法的未来发展,从而降低转移趋势。
4 甲状腺激素
甲状腺激素对大多数人体组织的生长、发育、基础代谢和生理功能都具有重要的生物学作用。甲状腺激素在体外具有促肿瘤作用,它们能以类似雌激素的方式刺激乳腺细胞的生长[27-28]。在啮齿动物模型中,它们刺激肿瘤的生长和癌细胞的转移扩散,反之,诱发甲状腺功能的减退则对BC和其他类型的癌症均有利。Søgaard等[29]进行了一项基于人群的队列研究,纳入了61 873例甲状腺功能低下的女性和80 343例甲状腺功能亢进的女性,发现相比一般人群,甲状腺功能亢进的女性患BC的风险更大,甲状腺功能低下的女性BC的发病率略有降低。Moretto等[30]使用超过生理剂量的T3处理MCF7乳腺癌细胞后,发现缺氧诱导因子1(hypoxia inducing factor 1, HIF-1)和转化生长因子α(transforming growth factor alpha, TGF-α)的表达增加,而这一现象是由T3激活PI3K所介导的,HIF-1和TGF-α的过表达与乳腺癌的侵袭性和恶性进展高度相关。除此之外,T4也被证明是体外乳腺癌细胞的增殖因子,可以促进核雌激素受体-α依赖性乳腺癌细胞的增殖,这种作用涉及MAPK依赖性蛋白激酶和特定丝氨酸磷酸化对ERα的激活。在缺乏雌激素受体的乳腺癌细胞中,甲状腺激素的促增殖作用则是由肿瘤细胞表达的整联蛋白αvβ3的胞外结构域上的甲状腺激素受体介导的。因此,无论ER存在与否,甲状腺激素都能够通过两种机制支持乳腺癌细胞的增殖[31]。关于甲状腺激素和BC之间联系的观点尚未统一,并且缺乏确凿的证据和大样本量的数据验证,因此,需要进一步研究以调查甲状腺激素与BC之间的确切联系。
甲状腺激素的合成与分泌主要受促甲状腺激素(thyroid stimulating hormone, TSH)调控,TSH通过与TSH受体结合以增强甲状腺激素合成的各个阶段。目前,研究人员发现与普通人群对照,DTC患者的血清TSH水平显著升高,且TSH的表达与肿瘤的侵略性相关。进一步的亚组分析显示,血清TSH在正常水平范围内,且超声检查未探及结节的人群患DTC风险也将随TSH的水平升高而升高[32],有关TSH与甲状腺癌的具体发病机制仍需要进一步研究。
5 甲状腺激素受体
甲状腺激素受体(thyroid hormone receptors, TRs)属于核激素受体家族,包括甲状腺激素受体α(TRα)和甲状腺激素受体β(TRβ),Heublein等[33]的研究证明,TRs可能与BRCA1相关BC患者的预后有关。TRβ阳性可能与BC患者的五年或总生存率正相关,而TRα具有相反的作用。在部分肿瘤中,经常可以观察到TR丢失的现象。根据先前的研究我们可以得知,TRβ在肿瘤的发生与发展中具有抑制作用,它可以通过抑制细胞外信号调节激酶和PI3K信号通路的激活来抑制甲状腺癌和乳腺癌细胞的增殖并促进细胞凋亡[34]。此外,TRβ还可以特异性结合甲状腺激素反应元件从而抑制癌基因RUNX2的转录活性[35],而RUNX2作为肿瘤的促进剂在甲状腺癌和乳腺癌中都发生异常表达。2015年,Park等[36]表示,TRβ羧基末端的移码突变PV,表现出了致癌活性。除此之外,TRβ1C末端的螺旋11和12的一些其他序列,如Mkar、Mdbs和AM的突变,也表现出对肿瘤的驱动作用,增强了细胞增殖和侵袭性并抑制凋亡。部分国内的研究也显示,TRβ1的表达降低和突变与中国人群BC的发生和发展有关[37]。因此,TR的功能障碍可能是导致BC与TC并发的因素之一。
6 甲状腺过氧化物酶自身抗体
研究人员发现将TC与BC并发的患者与仅TC组相比,并发组中抗甲状腺过氧化物酶自身抗体(antithyroid peroxidase autoantibody, TPOAb)的平均值较高,且BC患者的血清TPOAb水平明显高于健康人群。因此我们猜测乳腺癌和甲状腺癌之间存在共同的抗原介导免疫反应。众所周知,癌症和自身免疫反应均由免疫系统的异常所导致,一方面无法杀死肿瘤细胞,另一方面又损伤自身组织。因此,癌症和自身免疫可以通过双向免疫联系起来。Muller等[38]研究表明40%~45%的TPOAb与乳过氧化物酶(lactoperoxidase, LPO)有交叉反应,且LPO与甲状腺过氧化物酶(thyroid peroxidase, TPO)的同源性为46%,但大多数TPOAb都直接朝向TPO的免疫优势区域,并且这些似乎不会与不同的过氧化物酶发生交叉反应,因此,可能存在不同类型的循环TPOAb,其中一些与其他过氧化物酶(包括LPO)发生交叉反应,这解释了TPOAb在BC患者中更为普遍的原因。此外,LPO能与雌激素发生反应,将他们氧化为自由基,随后自由基与DNA相互作用,从而在乳腺癌的致病过程中发挥作用。
甲状腺过氧化物酶(thyroid peroxidase, TPO)不仅在甲状腺细胞中表达,还在BC和乳腺肿瘤周围组织中表达,但其水平远低于甲状腺细胞。即便如此,TPO表达水平也能够触发免疫反应,这可能为BC中TPOAb水平升高提供了另一种解释。虽然TPO在肿瘤周围的乳腺组织中也表达,但是TPOAb水平仅在BC中增加,因此研究人员推测了几种触发TPO免疫反应的机制,TPO和LPO都具有酶活性,可以诱导乳腺组织中的氧化应激并参与BC的发病机制。BC中存在的组织性微环境改变可能会改变LPO和(或)TPO的表达和抗原特性,并促进自身免疫[39]。同时,慢性和失调性炎性反应的存在与BC风险增加有关[40]。
7 其他
BC与TC的并发除受到内在的基因、激素、免疫的影响,还存在外在的物理、化学等因素影响。电离辐射是BC和TC公认的危险因素,1930—1940年,对于部分良、恶性疾病进行外照射是常用的治疗方法,如对痤疮的头颈部照射、对胸腺肿大的胸部照射以及对儿童期霍奇金淋巴瘤的放射治疗,接受这些治疗的人群在数年后,BC和TC的患病率显著升高。在切尔诺贝利、广岛和长崎等事故中,暴露于电离辐射的受害者患上BC和TC的概率也明显升高,这可能是由于辐射导致DNA损伤修复机制发生异常。另外,由生活方式导致的肥胖也被认作是BC与TC同时发生的危险因素,其在BC发展中的作用早已为人所知。而最近Schmid等[41]通过研究后发现肥胖与上皮来源的甲状腺癌发展高度相关,但与甲状腺C细胞来源的肿瘤没有明显关联。乳腺癌合并甲状腺癌的潜在机制,见表 1。
表 1 乳腺癌合并甲状腺癌的潜在机制Table 1 Underlying mechanism of breast cancer combined with thyroid cancer
8 结语
综上所述,原发性乳腺癌合并甲状腺癌可能与共同的致癌基因、内分泌激素的相互作用以及电离辐射等环境因素有关,因此,在对原发癌症诊断和随访的过程中,不仅要注意转移癌,还应对第二原发癌保持警惕。对于临床医生来说,准确的诊断是一项非常困难但很重要的任务,他们需要区分第二原发癌和由原发肿瘤转移而来的病灶,目前超声引导细针穿刺组织学活检是明确诊断的首选方法。在治疗过程中,临床医生应根据两者并发的致病因素,提出癌症预防和治疗的新靶标,并制定更完善的治疗方案,例如针对乳腺癌中钠碘同向转运体的放射性碘治疗、甲状腺癌的抗雌激素治疗等。虽然这些方法仍需进一步探索和改进,但其未来的应用前景将十分广阔。
Competing interests: The authors declare that they have no competing interests.作者贡献:马骁:数据收集分析、论文设计和撰写刘万红:论文选题及修改 -
表 1 未明确分型的11株HAdV病毒株信息
Table 1 Information of 11 unverified HAdV strains

表 2 MH634393、KF302429与D型HAdV基因组多序列比对结果
Table 2 Multiple sequence alignment results among MH634393, KF302429 and HAdV-D genome

表 3 MK570618与C型HAdV基因组序列多序列比对结果
Table 3 Multiple sequence alignment results among MK570618 and HAdV-C genome

表 4 MH634393、KF302429与B型HAdV基因组多序列比对结果
Table 4 Multiple sequence alignment results among MH634393, KF302429 and HAdV-B genome

表 5 MK570619与F型HAdV基因组多序列比对结果
Table 5 Multiple sequence alignment results among MK570619 and HAdV-F genome

表 6 未明确分型HAdV序列类型多序列比对结果
Table 6 Multiple sequence alignment results of unverified HAdV genome

-
[1] Mennechet FJD, Paris O, Ouoba AR, et al. A review of 65 years of human adenovirus seroprevalence[J]. Expert Rev Vaccines, 2019, 18(6): 597-613. doi: 10.1080/14760584.2019.1588113
[2] Lynch 3rd JP, Kajon AE. Adenovirus: Epidemiology, Global Spread of Novel Serotypes, and Advances in Treatment and Prevention[J]. Semin Respir Crit Care Med, 2016, 37(4): 586-602. doi: 10.1055/s-0036-1584923
[3] Chartier C, Degryse E, Gantzer M, et al. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli[J]. J Virol, 1996, 70(7): 4805-4810. doi: 10.1128/jvi.70.7.4805-4810.1996
[4] Xing L, Tikoo SK. Promoter activity of left inverted terminal repeat and downstream sequences of porcine adenovirus type 3[J]. Virus Res, 2005, 109(1): 51-58. doi: 10.1016/j.virusres.2004.10.006
[5] Kelly E, Russell SJ. History of oncolytic viruses: genesis to genetic engineering[J]. Mol Ther, 2007, 15(4): 651-659. doi: 10.1038/sj.mt.6300108
[6] Twumasi-Boateng K, Pettigrew JL, Kwok YYE, et al. Oncolytic viruses as engineering platforms for combination immunotherapy[J]. Nat Rev Cancer, 2018, 18(7): 419-432. doi: 10.1038/s41568-018-0009-4
[7] Vogels R, Zuijdeest D, van Rijnsoever R, et al. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: efficient human cell infection and bypass of preexisting adenovirus immunity[J]. J Virol, 2003, 77(15): 8263-8271. doi: 10.1128/JVI.77.15.8263-8271.2003
[8] Bishop MW, Janeway KA, Gorlick R. Future directions in the treatment of osteosarcoma[J]. Curr Opin Pediatr, 2016, 28(1): 26-33. doi: 10.1097/MOP.0000000000000298
[9] Baker AT, Aguirre-Hernández C, Halldén G, et al. Designer Oncolytic Adenovirus: Coming of Age[J]. Cancers (Basel), 2018, 10(6): 201. doi: 10.3390/cancers10060201
[10] Burland TG. DNASTAR's Lasergene sequence analysis software[J]. Methods Mol Biol, 2000, 132: 71-91.
[11] Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets[J]. Mol Biol Evol, 2016, 33(7): 1870-1874. doi: 10.1093/molbev/msw054
[12] Scott MK, Chommanard C, Lu X, et al. Human Adenovirus Associated with Severe Respiratory Infection, Oregon, USA, 2013-2014[J]. Emerg Infect Dis, 2016, 22(6): 1044-1051. doi: 10.3201/eid2206.151898
[13] Arnberg N. Adenovirus receptors: implications for targeting of viral vectors[J]. Trends Pharmacol Sci, 2012, 33(8): 442-448. doi: 10.1016/j.tips.2012.04.005
[14] de Graaf JF, de Vor L, Fouchier RAM, et al. Armed oncolytic viruses: A kick-start for anti-tumor immunity[J]. Cytokine Growth Factor Rev, 2018, 41: 28-39. doi: 10.1016/j.cytogfr.2018.03.006
[15] Nguyen TV, Crosby CM, Heller GJ, et al. Oncolytic adenovirus Ad657 for systemic virotherapy against prostate cancer[J]. Oncolytic Virother, 2018, 7: 43-51. doi: 10.2147/OV.S155946
-
期刊类型引用(5)
1. 安俊达,李玉双. 基于超声图像评估甲状腺和乳腺病变的通用计算方法. 燕山大学学报. 2024(01): 86-94 .  百度学术
百度学术
2. 杨茜雯,宋博,高婷婷,薛无瑕,李立坤,纪永章. 2021—2022年某三甲医院体检人群重要异常结果分布特征分析. 健康体检与管理. 2024(02): 174-177+202 .  百度学术
百度学术
3. 杨倩倩,夏源,张云飞,杨桂云,郭文佳,张秀华. 基于列线图的女性乳腺癌患者再发第二原发性甲状腺癌风险预测模型的构建和验证. 中国医药导报. 2024(24): 117-124 .  百度学术
百度学术
4. 阙厦丹,陈敦雁. 乳腺良性肿瘤和恶性肿瘤与自身免疫性甲状腺疾病的关系. 慢性病学杂志. 2023(05): 755-757 .  百度学术
百度学术
5. 王婷. 中西医结合治疗乳腺癌术后甲状腺结节的临床疗效. 世界复合医学. 2022(04): 109-111+116 .  百度学术
百度学术
其他类型引用(1)




 下载:
下载: